Visualization Package for Evolutionary Dynamics from Sequence and Model Data. biorxiv pre-print here.
Users have options ot install EvoFreq depending on if they want to use 'devtools'. Below we show two popular methods for installation. You will need a fully operational install of R (≥3.5.0) and, optionally, RStudio.
With devtools:
install.packages('devtools')
library(devtools)
install_github('MathOnco/EvoFreq')From source:
# clone the repository and pass 'install.packages' the path to the cloned evofreq repository
install.packages("./EvoFreq/", repos = NULL, type="source")
library("EvoFreq")
# To uninstall this if it failed, use:
detach("package:EvoFreq", unload = TRUE)
remove.packages("EvoFreq")EvoFreq has two primary functions: get_evofreq() and the final rendering of these frequencies using plot_evofreq(). To quckly see all the functions within EvoFreq use help(package="EvoFreq"). The quickest way to see an EvoFreq plot is to use:
library(EvoFreq)
data("example.easy.wide") # load example data
freq_frame <- get_evofreq(example.easy.wide[,seq(3,10)], example.easy.wide$clones, example.easy.wide$parents, clone_cmap = "magma") # Get freq_frame, a properly structured data.frame
evo_freq_p <- plot_evofreq(freq_frame) # Get an EvoFreq plotBelow you will find more useful tools, information, and features.
A goal of EvoFreq was to increase the flexibility in the input data. To this end we employ functions to use both long and wide dataframes.
# Note: this can be copy and pasted after installed
data("example.easy.wide") # Load a simple Data Frame example
str(example.easy.wide) # Inspect the data structure
# 'data.frame': 8 obs. of 10 variables:
# $ parents: num 0 1 1 3 1 5 5 5
# $ clones : num 1 2 3 4 5 6 7 8
# $ 1 : num 1 0 0 0 0 0 0 0
# $ 2 : num 100 5 0 0 0 0 0 0
# $ 3 : num 200 100 5 0 0 0 0 0
# $ 4 : num 400 5 100 1 1 0 0 0
# $ 5 : num 0 0 200 100 100 1 0 1
# $ 6 : num 0 0 200 125 200 10 1 15
# $ 7 : num 0 0 300 200 300 20 10 25
# $ 8 : num 0 0 300 300 300 25 25 100
# You have A column of parents and a column of clones then you have a column for each of the timepoints with sizes for that clone.
# Then get the frequency data. (Use ?get_evofreq for options)
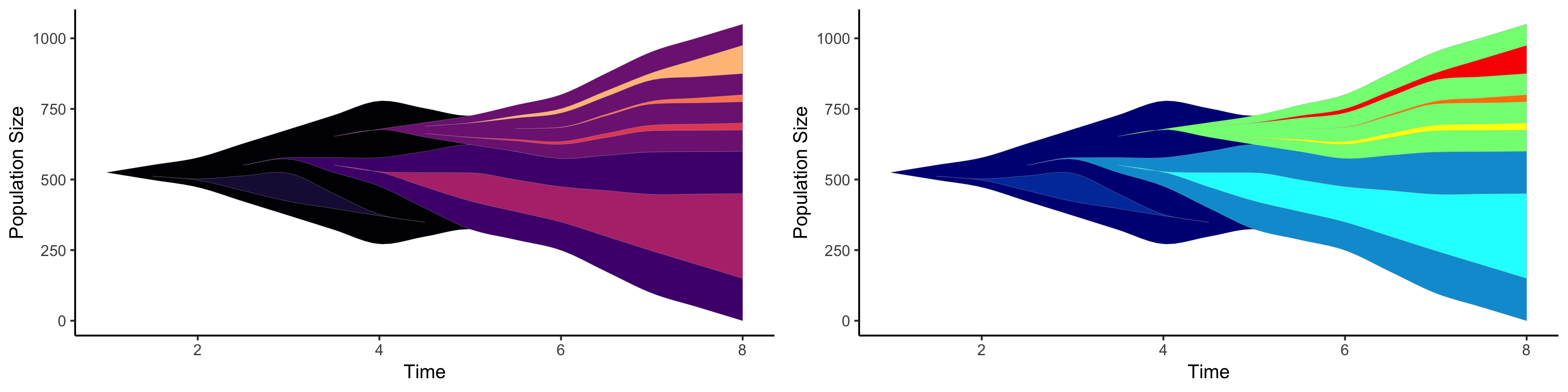
freq_frame <- get_evofreq(example.easy.wide[,seq(3,10)], example.easy.wide$clones, example.easy.wide$parents, clone_cmap = "magma")
# Create the plot (shown on the left below)
evo_freq_p <- plot_evofreq(freq_frame)
print(evo_freq_p)
# We can also choose to update the colors or do this during the first creation. (shown on the right below)
clone_dynamics_df_jet <- update_colors(freq_frame, example.easy.wide$clones, clone_cmap = "jet")
evo_freq_p_jet <- plot_evofreq(clone_dynamics_df_jet)
print(evo_freq_p_jet)# Note: this can be copy and pasted after installed
# In this example you have two files. One is the edge list of clones and their parents
# The other file is the sizes over time for those clones
# examine the structures to see how to format
# example.easy.long.edges now has an attribute column as well
data("example.easy.long.edges")
data("example.easy.long.sizes")
# Use the long_to_wide_size_df function to get the right data structure.
wide_df <- long_to_wide_freq_ready(long_pop_sizes_df = example.easy.long.sizes, time_col_name = "Time", clone_col_name = "clone", parent_col_name = "parent", size_col_name = "Size", edges_df = example.easy.long.edges)
clones <- wide_df$clones
parents <- wide_df$parents
size_df <- wide_df$wide_size_df
freq_frame <- get_evofreq(size_df, clones, parents, clone_cmap = "inferno")
evo_freq_p <- plot_evofreq(freq_frame)
evo_freq_p
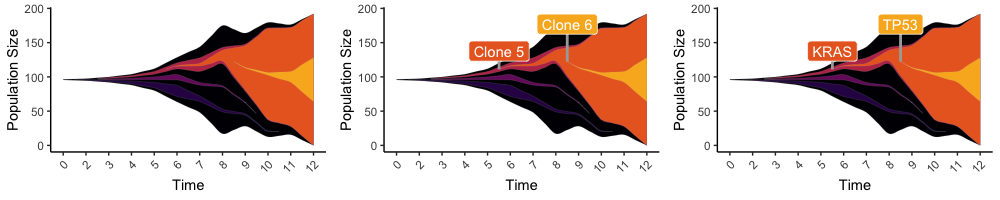
# Add custom ggplot features
evo_freq_labeled_p <- get_evofreq_labels(freq_frame, clone_list=c(5,6), extant_only=F, evofreq_plot = evo_freq_p, apply_labels=T)
evo_freq_labeled_p
# Add custom labels instead of defaults
evo_freq_labeled_p_custom <- get_evofreq_labels(freq_frame, clone_list=c(5,6), custom_label_text = c("KRAS","TP53"), extant_only=F, evofreq_plot = evo_freq_p, apply_labels=T)
evo_freq_labeled_p_custom
library(gridExtra)
grid.arrange(evo_freq_p,evo_freq_labeled_p,evo_freq_labeled_p_custom, nrow=1)EvoFreq has necessary functions for visualizing PhyloWGS and CALDER outputs. Similar to other tools, CloneEvol outputs are already compatible.
# PhyloWGS
phylowgs_output="run_name.summ.json"
# Return one or all using parsing options. Use "?parse_phylowgs" for help
tree_data <- parse_phylowgs(json_file=phylowgs_output)
#EvoFreqPlots
pdf('./evofreqs.pdf', width=8, height=4, onefile = T)
for (i in 1:length(f$all)){
clone_dynamics_df <- get_evofreq(tree_data[[i]][,c(5,length(colnames(tree_data)))], clones=tree_data[[i]]$clone, parents=tree_data[[i]]$parent, clone_cmap = "jet")
p <- plot_evofreq(evofreq_df)
print(p)
}
dev.off()
# CALDER
theFile <- "SA501_tree1.dot"
theSoln <- "SA501_soln1.csv"
calder.data <- parse_calder(theSoln, theFile)
### Use the long_to_wide_size_df function to get the right data structure.
wide_df <- long_to_wide_size_df(long_pop_sizes_df = calder.data$sizeDf,
edges_df = calder.data$edges,
time_col_name = "time",
clone_col_name = "clone",
parent_col_name = "parent",
size_col_name = "size",
fill_gaps_in_size = T
)
clones <- as.character(wide_df$clones)
parents <- as.character(wide_df$parents)
size_df <- wide_df$wide_size_df
clone_dynamics_df <- get_evofreq(size_df, clones, parents, clone_cmap = "jet", data_type = "size", threshold=0, test_links = T, add_origin = T, interp_method = "bezier")
plot_evofreq(clone_dynamics_df)One of the most powerful features is the endless additions that can be added to each plot. Any ggplot2 function can be added to the frequency dynamics plots.
For a full list of the different functions and example datasets please use help(package="EvoFreq").
HAL now provides functionality that makes visualizing model data with EvoFreq easy. A great blog post illustrates how to do this found here.
These papers have used EvoFreq for their publication ready images.
- Chandler D. Gatenbee, Ann-Marie Baker, Ryan O. Schenck, Margarida P. Neves, Sara Yakub Hasan, Pierre Martinez, William CH Cross, Marnix Jansen, Manuel Rodriguez-Justo, Andrea Sottoriva, Simon Leedham, Mark Robertson-Tessi, Trevor A. Graham, Alexander R.A. Anderson. Niche engineering drives early passage through an immune bottleneck in progression to colorectal cancer. 2019. bioRxiv.
- Ryan O Schenck, Eunung Kim, Rafael Bravo, Jeffrey West, Simon Leedham, Darryl Shibata, Alexander R.A. Anderson. Clonal Architecture of the Epidermis: Homeostasis Limits Keratinocyte Evolution. 2019. bioRxiv.
- Jeffrey West, Ryan Schenck, Chandler Gatenbee, Mark Robertson-Tessi, Alexander RA Anderson. Tissue structure accelerates evolution: premalignant sweeps precede neutral expansion. 2019. bioRxiv.
- Jeffrey West, Li You, Joel Brown, Paul K. Newton, Alexander R. A. Anderson. Towards multi-drug adaptive therapy. 2019. bioRxiv.