Automatic walking strides segmentation from wrist-worn sensor accelerometry data collected in the free-living
- Accelerometry data sample
- Segment individual walking strides
- Estimation of walking cadence (steps/min)
- Session info
- References
Below, we demonstrate our method for automatic walking strides segmentation from wrist-worn sensor accelerometry data collected in the free-living environment.
This repository accompanies “Estimation of free-living walking cadence
from wrist-worn sensor accelerometry data and its association with SF-36
quality of life scores” manuscript ([1]) in which the method is
proposed. The method is implemented as the segmentWalking() function
in adept R package (CRAN
index).
Below, we demonstrate the method with the use of independent data sample
(not used in the manuscript study).
The directory data/ contains data files:
acc_raw_left_wrist.rdsacc_raw_right_wrist.rds
which are a sample of raw accelerometry data collected consecutively for
2 days, starting and ending at midnight of a week day. Data were
collected at the sampling frequency of 100 Hz with two ActiGraph GT9X
Link sensors located at the left wrist and right wrist, respectively.
One data observation includes (x,y,z) values collected from each of
the three axes at the same time. At the sampling frequency of 100 Hz,
there were 100 observations collected per second (17,280,000
observations per 2 days) for each of the two sensors.
Below, we read the data and plot a few first and last observations from left wrist data set.
library(tidyverse)
library(lubridate)
library(adeptdata) # for stride pattern templates data
library(adept)
# read raw accelerometry data from left wrist, right wrist
dat_lw <- readRDS(paste0(here::here(), "/data/acc_raw_left_wrist.rds")) %>% as_tibble()
dat_rw <- readRDS(paste0(here::here(), "/data/acc_raw_right_wrist.rds")) %>% as_tibble()
# define data collection POSIXct (used *fake* day date of Feb 1, 2021)
obs_datetime_seq <- seq(ymd_hms("2021-02-01 00:00:00"), by = 0.01, length.out = nrow(dat_lw))
dat_lw <- mutate(dat_lw, obs_datetime = obs_datetime_seq, .before = "x")
dat_rw <- mutate(dat_rw, obs_datetime = obs_datetime_seq, .before = "x")rbind(head(dat_lw, 3), tail(dat_lw, 3))
#> # A tibble: 6 x 6
#> loc_id obs_idx obs_datetime x y z
#> <chr> <int> <dttm> <dbl> <dbl> <dbl>
#> 1 left_wrist 1 2021-02-01 00:00:00 1 0.176 0.016
#> 2 left_wrist 2 2021-02-01 00:00:00 1 0.176 0.016
#> 3 left_wrist 3 2021-02-01 00:00:00 1 0.176 0.016
#> 4 left_wrist 17279998 2021-02-02 23:59:59 -0.977 -0.078 -0.188
#> 5 left_wrist 17279999 2021-02-02 23:59:59 -0.977 -0.078 -0.188
#> 6 left_wrist 17280000 2021-02-02 23:59:59 -0.977 -0.078 -0.188Data were collected from a healthy 28F person.
(Click to see IRB note.)
The raw accelerometry data in `data/` package were collected from sensors worn by Marta Karas, an author of this repository. The IRB Office Determination Request Form for Primary (New) Data Collection request form was submitted in regard to the collection and further publishing of these data. Based on preliminary review of the request form submitted, it was determined that the data collection and further data publishing activity described in the determination request does not qualify as human subjects research as defined by DHHS regulations 45 CFR 46.102, and does not require IRB oversight.One way to visualize raw accelerometry data is to plot it as a
three-dimensional time-series (x,y,z). Here, we plot data from three
different time frames, each of 4 seconds length, simultaneously for data
collected at left wrist and right wrist.
(Click to see the code.)
# define time frame start values for data subset
t1 <- ymd_hms("2021-02-01 09:57:11")
t2 <- ymd_hms("2021-02-01 09:59:41")
t3 <- ymd_hms("2021-02-01 10:03:47")
# combine data from two sensors, subset to keep only selected time windows
dat <- rbind(dat_lw, dat_rw)
dat_sub <- dat %>%
filter((obs_datetime >= t1 & obs_datetime < t1 + as.period(4, "seconds")) |
(obs_datetime >= t2 & obs_datetime < t2 + as.period(4, "seconds")) |
(obs_datetime >= t3 & obs_datetime < t3 + as.period(4, "seconds")) ) %>%
mutate(dt_floor = paste0("time frame start: ",
floor_date(obs_datetime, unit = "minutes")))
# plot (x,y,z) values
dat_sub %>%
select(-obs_idx) %>%
pivot_longer(cols = -c(dt_floor, obs_datetime, loc_id)) %>%
ggplot(aes(x = obs_datetime, y = value, color = name)) +
geom_line(size = 0.3) +
facet_grid(loc_id ~ dt_floor, scales = "free_x") +
theme_minimal(base_size = 10) +
labs(x = "Time [s]",
y = "Acceleration [g]",
color = "Accelerometer axis of measurement: ") +
theme(legend.position = "top")Vector magnitude (VM) is often used to reduce the dimensionality of
accelerometry time-series (x,y,z). VM is computed as
vm = sqrt(x^2 + y^2 + z^2) at each time point resulting in 1- instead
of 3-dimensional time-series.
(Click to see the code.)
# plot vector magnitude values
dat_sub %>%
mutate(vm = sqrt(x^2 + y^2 + z^2)) %>%
ggplot(aes(x = obs_datetime, y = vm)) +
geom_line(size = 0.3) +
facet_grid(loc_id ~ dt_floor, scales = "free_x") +
theme_minimal(base_size = 10) +
labs(x = "Time [s]", y = "VM") We use segmentWalking() function in adept R package to automatically
segment individual walking strides from the raw accelerometry data
collected with a wrist-worn sensor.
First, the function first uses Adaptive Empirical Pattern Transformation (ADEPT) method (see ADEPT manuscript [2]) to segment walking stride patterns. In short, ADEPT uses a predefined template and detects its repetitions in data by maximizing the local correlation between scale-transformed template’s versions and the observed data at every time point. Multiple baseline templates can be used simultaneously to best fit various shapes of strides. The ADEPT result describes each segmented stride by its:
-
- start time,
-
- duration (seconds),
-
- correlation with best-fit template.
Second, the function filters the patterns segmented from the data to keep those which plausibly and with high specificity correspond to walking activity. Methodological details are reported in the method manuscript ([1]).
Below, we use three wrist-specific stride templates publicly available in adeptdata R package.
(Click to see the code.)
# pull 3 x 200 matrix with 3 distinct stride pattern templates
# (attached in adeptdata R package)
templates_mat <- stride_template$left_wrist[[3]]
plt_df <-
templates_mat %>%
as.data.frame() %>%
rename_all(~as.character(seq(0, 1, length.out = ncol(templates_mat)))) %>%
mutate(template_idx = row_number()) %>%
pivot_longer(cols = -template_idx) %>%
mutate(name = as.numeric(name),
template_idx = factor(template_idx, labels = paste0("Template ", 1 : 3)))
ggplot(plt_df, aes(x = name, y = value)) +
geom_line() +
facet_wrap(~ template_idx) +
theme_minimal(base_size = 10) +
labs(x = "Templaste phase", y = "VM") +
labs(x = "Time [s]", y = "VM") We use segmentWalking() method from adept R package to segment
walking separately from data from (a) left wrist, (b) right-wrist worn
sensor. Default algorithm parameters are optimized for a wrist-worn
sensor.
(Click to see the code – part 1.)
# convert templates matrix to templates list
templates_list <- list(templates_mat[1, ], templates_mat[2, ], templates_mat[3, ])
# segment walking from data collected with a left wrist-worn sensor
t1 <- Sys.time()
out_lw <- segmentWalking(
xyz = dat_lw %>% select(all_of(c("x", "y", "z"))),
xyz.fs = 100,
template = templates_list,
run.parallel = TRUE,
run.parallel.cores = 8)
Sys.time() - t1
# Time difference of 10.45637 mins
# segment walking from data collected with a right wrist-worn sensor
t1 <- Sys.time()
out_rw <- segmentWalking(
xyz = dat_rw %>% select(all_of(c("x", "y", "z"))),
xyz.fs = 100,
template = templates_list,
run.parallel = TRUE,
run.parallel.cores = 8)
Sys.time() - t1
# Time difference of 5.958216 mins
# save precomputed results for faster README compilation
saveRDS(out_lw, paste0(here::here(), "/data/out_lw.rds"))
saveRDS(out_rw, paste0(here::here(), "/data/out_rw.rds"))(Click to see the code – part 2.)
# read precomputed results for faster README compilation
out_lw <- readRDS(paste0(here::here(), "/data/out_lw.rds"))
out_rw <- readRDS(paste0(here::here(), "/data/out_rw.rds"))
# filter output to keep rows corresponding to identified walking
out_lw_1 <- out_lw %>% filter(is_walking_i == 1)
out_rw_1 <- out_rw %>% filter(is_walking_i == 1)
head(out_lw_1)
#> tau_i T_i sim_i template_i is_walking_i
#> 1 3595614 112 0.8986153 NA 1
#> 2 3595725 111 0.9564651 NA 1
#> 3 3595835 122 0.9417612 NA 1
#> 4 3598471 113 0.9049312 NA 1
#> 5 3598583 110 0.9398946 NA 1
#> 6 3598692 131 0.8926561 NA 1Below, we visualize first K=50 strides segmented from (a) left wrist, (b) right-wrist worn sensor.
(Click to see the code.)
# get VM from raw data frames
dat_lw_vm <- sqrt(dat_lw$x^2 + dat_lw$y^2 + dat_lw$z^2)
dat_rw_vm <- sqrt(dat_rw$x^2 + dat_rw$y^2 + dat_rw$z^2)
K <- 50
T_i_max <- max(c(out_lw_1$T_i, out_rw_1$T_i))
stride_dat <- numeric()
stride_id <- numeric()
stride_loc <- numeric()
for (k in 1 : K){ # k <- 1
# left wrist data
dat_idx_k <- (out_lw_1[k, "tau_i"]) : (out_lw_1[k, "tau_i"] + out_lw_1[k, "T_i"] - 1)
dat_k <- dat_lw_vm[dat_idx_k]
stride_dat <- c(stride_dat, dat_k)
stride_id <- c(stride_id, rep(k, length(dat_k)))
stride_loc <- c(stride_loc, rep("left_wrist", length(dat_k)))
# right wrist data
dat_idx_k <- (out_rw_1[k, "tau_i"]) : (out_rw_1[k, "tau_i"] + out_rw_1[k, "T_i"] - 1)
dat_k <- dat_rw_vm[dat_idx_k]
stride_dat <- c(stride_dat, dat_k)
stride_id <- c(stride_id, rep(k, length(dat_k)))
stride_loc <- c(stride_loc, rep("right_wrist", length(dat_k)))
}
# define plot data frame
plt_df <-
data.frame(stride_dat, stride_id, stride_loc) %>%
group_by(stride_id, stride_loc) %>%
mutate(stride_duration = row_number() - 1,
stride_duration = stride_duration / 100) %>%
ungroup()
# point-wise aggregate of strides
plt_df_agg <-
plt_df %>%
group_by(stride_loc, stride_duration) %>%
summarise(stride_dat = mean(stride_dat)) %>%
ungroup()
ggplot(plt_df, aes(x = stride_duration, y = stride_dat, group = stride_id)) +
geom_line(alpha = 0.2) +
facet_wrap(~ stride_loc) +
theme_minimal(base_size = 10) +
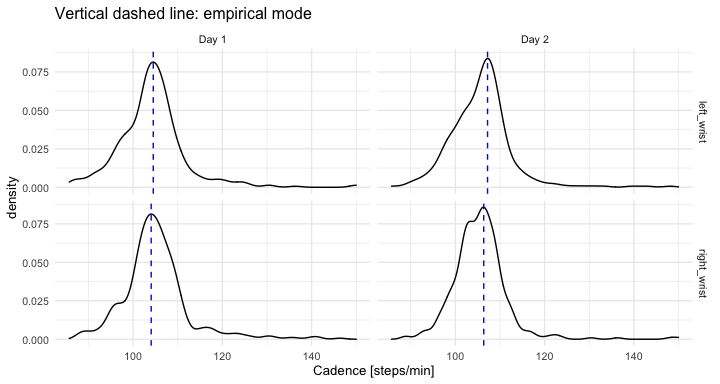
labs(x = "Stride duration [s]", y = "VM") The data used in this tutorial consists of two days of continuous monitoring. We use
segmented strides to estimate the walking cadence (number of steps per minute) at every time of the day when walking was identified. Next, we compute the mode of all estimated cadences during the day was computed – a daily cadence measurement.
(Click to see the code.)
# function to compute mode
estimate_mode <- function(x) {
d <- density(x)
d$x[which.max(d$y)]
}
# data collection frequency
hz_val <- 100
# combine result from walking segmentation from left and right wrist
out_comb_1 <- rbind(
out_lw_1 %>% mutate(loc_id = "left_wrist"),
out_rw_1 %>% mutate(loc_id = "right_wrist")
) %>%
mutate(
# define stride duration (in seconds) as number of observations in stride
# (stride := two subsequent steps)
# over data frequency (number of observations per 1 second)
dur_i = T_i / hz_val,
# define cadence as number of steps per 1 minute
cadence_i = (2/dur_i) * 60,
# define collection day "ID"
day_id = ceiling(tau_i / (hz_val * 60 * 60 * 24)),
day_id = factor(day_id, levels = c(1,2), labels = paste0("Day ", 1:2))
)
# aggregate the cadence measurements by computing empirical mode
# (1 value per day)
out_comb_1_mode <-
out_comb_1 %>%
group_by(loc_id, day_id) %>%
summarise(stride_duration_mode = estimate_mode(dur_i),
cadence_mode = estimate_mode(cadence_i)) %>%
arrange(day_id, loc_id) %>%
as.data.frame()
ggplot(out_comb_1, aes(x = cadence_i)) +
geom_density() +
facet_grid(loc_id ~ day_id) +
geom_vline(data = out_comb_1_mode, aes(xintercept = cadence_mode),
linetype = 2, color = "blue") +
theme_minimal(base_size = 10) +
labs(x = "Cadence [steps/min]", title = "Vertical dashed line: empirical mode")The table summarizes empirical mode of stride duration [s] and cadence [steps/min]:
(Click to see the code.)
print(out_comb_1_mode)
#> loc_id day_id dur_i_mode cadence_i_mode
#> 1 left_wrist Day 1 1.147018 1.741989
#> 2 right_wrist Day 1 1.150091 1.734214
#> 3 left_wrist Day 2 1.117782 1.786894
#> 4 right_wrist Day 2 1.128068 1.772624#> loc_id day_id dur_i_mode cadence_i_mode
#> 1 left_wrist Day 1 1.147018 1.741989
#> 2 right_wrist Day 1 1.150091 1.734214
#> 3 left_wrist Day 2 1.117782 1.786894
#> 4 right_wrist Day 2 1.128068 1.772624
Similarly, next, we compute the mode of all estimated cadences during the day-hour was computed. We visualize hourly cadence estimated for day-hours where the number of estimated walking strides is more or equal 10.
(Click to see the code.)
# compute hour ID
out_comb_1 <-
out_comb_1 %>%
mutate(
hour_id = floor(tau_i / (hz_val * 60 * 60)),
hour_id = hour_id %% 24
)
# aggregate the cadence measurements by computing empirical mode
# (1 value per day-hour)
out_comb_1_mode_hourly <-
out_comb_1 %>%
group_by(loc_id, day_id, hour_id) %>%
summarise(cadence_mode = estimate_mode(cadence_i),
cnt = n()) %>%
as.data.frame()
ggplot(out_comb_1_mode_hourly %>% filter(cnt > 10),
aes(x = hour_id, y = cadence_mode, group = 1)) +
geom_line(linetype = 2, size = 0.5, color = "grey") +
geom_point(aes(size = cnt, alpha = cnt)) +
facet_grid(loc_id ~ day_id) +
theme_minimal(base_size = 10) +
labs(x = "Hour of a day",
y = "Cadence [steps/min]",
alpha = "Segmented\nstrides\ncount",
size = "Segmented\nstrides\ncount") +
scale_x_continuous(limits = c(0, 23), breaks = seq(0, 23, by = 2))(Click to see session info.)
devtools::session_info()
#> ─ Session info ───────────────────────────────────────────────────────────────
#> setting value
#> version R version 4.0.3 (2020-10-10)
#> os macOS Catalina 10.15.7
#> system x86_64, darwin17.0
#> ui X11
#> language (EN)
#> collate en_US.UTF-8
#> ctype en_US.UTF-8
#> tz America/New_York
#> date 2021-03-07
#>
#> ─ Packages ───────────────────────────────────────────────────────────────────
#> package * version date lib source
#> adept * 1.2 2021-01-31 [1] local
#> adeptdata * 1.0.1 2019-03-30 [1] CRAN (R 4.0.2)
#> assertthat 0.2.1 2019-03-21 [1] CRAN (R 4.0.2)
#> backports 1.2.1 2020-12-09 [1] CRAN (R 4.0.2)
#> broom 0.7.5 2021-02-19 [1] CRAN (R 4.0.3)
#> cachem 1.0.4 2021-02-13 [1] CRAN (R 4.0.2)
#> callr 3.5.1 2020-10-13 [1] CRAN (R 4.0.2)
#> cellranger 1.1.0 2016-07-27 [1] CRAN (R 4.0.2)
#> cli 2.3.0 2021-01-31 [1] CRAN (R 4.0.3)
#> codetools 0.2-18 2020-11-04 [1] CRAN (R 4.0.2)
#> colorspace 2.0-0 2020-11-11 [1] CRAN (R 4.0.2)
#> crayon 1.4.1 2021-02-08 [1] CRAN (R 4.0.2)
#> data.table 1.14.0 2021-02-21 [1] CRAN (R 4.0.3)
#> DBI 1.1.1 2021-01-15 [1] CRAN (R 4.0.2)
#> dbplyr 2.1.0 2021-02-03 [1] CRAN (R 4.0.2)
#> desc 1.2.0 2018-05-01 [1] CRAN (R 4.0.2)
#> devtools 2.3.2 2020-09-18 [1] CRAN (R 4.0.2)
#> digest 0.6.27 2020-10-24 [1] CRAN (R 4.0.2)
#> dplyr * 1.0.4 2021-02-02 [1] CRAN (R 4.0.2)
#> dvmisc 1.1.4 2019-12-16 [1] CRAN (R 4.0.2)
#> ellipsis 0.3.1 2020-05-15 [1] CRAN (R 4.0.2)
#> evaluate 0.14 2019-05-28 [1] CRAN (R 4.0.1)
#> fansi 0.4.2 2021-01-15 [1] CRAN (R 4.0.2)
#> farver 2.0.3 2020-01-16 [1] CRAN (R 4.0.2)
#> fastmap 1.1.0 2021-01-25 [1] CRAN (R 4.0.2)
#> forcats * 0.5.1 2021-01-27 [1] CRAN (R 4.0.2)
#> fs 1.5.0 2020-07-31 [1] CRAN (R 4.0.2)
#> generics 0.1.0 2020-10-31 [1] CRAN (R 4.0.2)
#> ggplot2 * 3.3.3 2020-12-30 [1] CRAN (R 4.0.2)
#> glue 1.4.2 2020-08-27 [1] CRAN (R 4.0.2)
#> gtable 0.3.0 2019-03-25 [1] CRAN (R 4.0.2)
#> haven 2.3.1 2020-06-01 [1] CRAN (R 4.0.2)
#> highr 0.8 2019-03-20 [1] CRAN (R 4.0.2)
#> hms 1.0.0 2021-01-13 [1] CRAN (R 4.0.2)
#> htmltools 0.5.1.1 2021-01-22 [1] CRAN (R 4.0.2)
#> httr 1.4.2 2020-07-20 [1] CRAN (R 4.0.2)
#> jsonlite 1.7.2 2020-12-09 [1] CRAN (R 4.0.2)
#> knitr 1.31 2021-01-27 [1] CRAN (R 4.0.2)
#> labeling 0.4.2 2020-10-20 [1] CRAN (R 4.0.2)
#> lattice 0.20-41 2020-04-02 [1] CRAN (R 4.0.3)
#> lifecycle 1.0.0 2021-02-15 [1] CRAN (R 4.0.2)
#> lubridate * 1.7.9.2 2020-11-13 [1] CRAN (R 4.0.2)
#> magrittr 2.0.1 2020-11-17 [1] CRAN (R 4.0.2)
#> MASS 7.3-53.1 2021-02-12 [1] CRAN (R 4.0.2)
#> Matrix 1.3-2 2021-01-06 [1] CRAN (R 4.0.2)
#> memoise 2.0.0 2021-01-26 [1] CRAN (R 4.0.2)
#> mitools 2.4 2019-04-26 [1] CRAN (R 4.0.2)
#> modelr 0.1.8 2020-05-19 [1] CRAN (R 4.0.2)
#> munsell 0.5.0 2018-06-12 [1] CRAN (R 4.0.2)
#> mvtnorm 1.1-1 2020-06-09 [1] CRAN (R 4.0.2)
#> pillar 1.5.0 2021-02-22 [1] CRAN (R 4.0.3)
#> pkgbuild 1.2.0 2020-12-15 [1] CRAN (R 4.0.2)
#> pkgconfig 2.0.3 2019-09-22 [1] CRAN (R 4.0.2)
#> pkgload 1.1.0 2020-05-29 [1] CRAN (R 4.0.2)
#> pracma 2.3.3 2021-01-23 [1] CRAN (R 4.0.2)
#> prettyunits 1.1.1 2020-01-24 [1] CRAN (R 4.0.2)
#> processx 3.4.5 2020-11-30 [1] CRAN (R 4.0.2)
#> ps 1.5.0 2020-12-05 [1] CRAN (R 4.0.2)
#> purrr * 0.3.4 2020-04-17 [1] CRAN (R 4.0.2)
#> R6 2.5.0 2020-10-28 [1] CRAN (R 4.0.2)
#> rbenchmark 1.0.0 2012-08-30 [1] CRAN (R 4.0.2)
#> Rcpp 1.0.6 2021-01-15 [1] CRAN (R 4.0.2)
#> readr * 1.4.0 2020-10-05 [1] CRAN (R 4.0.2)
#> readxl 1.3.1 2019-03-13 [1] CRAN (R 4.0.2)
#> remotes 2.2.0 2020-07-21 [1] CRAN (R 4.0.2)
#> reprex 1.0.0 2021-01-27 [1] CRAN (R 4.0.2)
#> rlang 0.4.10 2020-12-30 [1] CRAN (R 4.0.2)
#> rmarkdown 2.7 2021-02-19 [1] CRAN (R 4.0.3)
#> rprojroot 2.0.2 2020-11-15 [1] CRAN (R 4.0.2)
#> rstudioapi 0.13 2020-11-12 [1] CRAN (R 4.0.2)
#> rvest 0.3.6 2020-07-25 [1] CRAN (R 4.0.2)
#> scales 1.1.1 2020-05-11 [1] CRAN (R 4.0.2)
#> sessioninfo 1.1.1 2018-11-05 [1] CRAN (R 4.0.2)
#> stringi 1.5.3 2020-09-09 [1] CRAN (R 4.0.2)
#> stringr * 1.4.0 2019-02-10 [1] CRAN (R 4.0.2)
#> survey 4.0 2020-04-03 [1] CRAN (R 4.0.2)
#> survival 3.2-7 2020-09-28 [1] CRAN (R 4.0.3)
#> tab 4.1.1 2019-06-17 [1] CRAN (R 4.0.2)
#> testthat 3.0.2 2021-02-14 [1] CRAN (R 4.0.2)
#> tibble * 3.0.6 2021-01-29 [1] CRAN (R 4.0.2)
#> tidyr * 1.1.2 2020-08-27 [1] CRAN (R 4.0.2)
#> tidyselect 1.1.0 2020-05-11 [1] CRAN (R 4.0.2)
#> tidyverse * 1.3.0 2019-11-21 [1] CRAN (R 4.0.2)
#> usethis 2.0.1 2021-02-10 [1] CRAN (R 4.0.2)
#> utf8 1.1.4 2018-05-24 [1] CRAN (R 4.0.2)
#> vctrs 0.3.6 2020-12-17 [1] CRAN (R 4.0.2)
#> withr 2.4.1 2021-01-26 [1] CRAN (R 4.0.2)
#> xfun 0.21 2021-02-10 [1] CRAN (R 4.0.2)
#> xml2 1.3.2 2020-04-23 [1] CRAN (R 4.0.2)
#> xtable 1.8-4 2019-04-21 [1] CRAN (R 4.0.2)
#> yaml 2.2.1 2020-02-01 [1] CRAN (R 4.0.2)
#>
#> [1] /Library/Frameworks/R.framework/Versions/4.0/Resources/library[1]: Karas, M., Urbanek, J.K., Illiano, V.P., Bogaarts, G., Crainiceanu, C.M., Dorn, J.F. (2021). Estimation of free-living walking cadence from wrist-worn sensor accelerometry data and its association with SF-36 quality of life scores, Submitted.
[2]: Karas, M., Straczkiewicz, M., Fadel, W., Harezlak, J., Crainiceanu, C.M., Urbanek, J.K. (2018). Adaptive empirical pattern transformation (ADEPT) with application to walking stride segmentation, Biostatistics, kxz033.