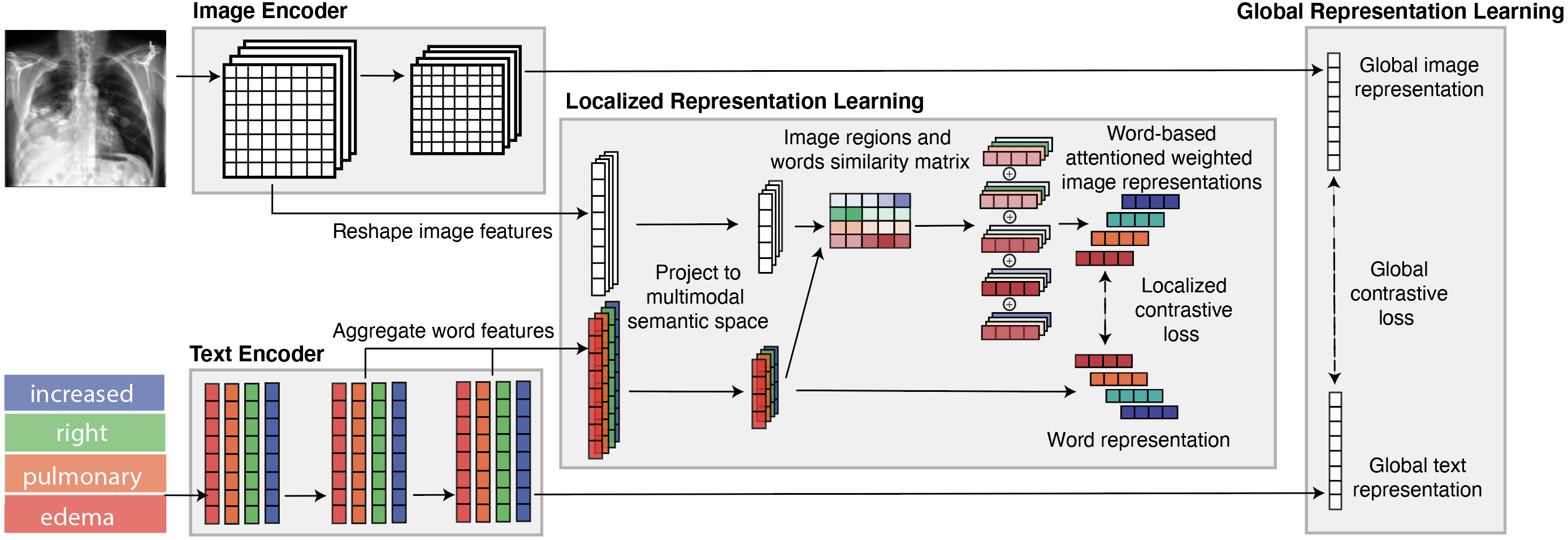
GLoRIA: A Multimodal Global-Local Representation Learning Framework for Label-efficient Medical Image Recognition
GLoRIA (Global-Local Representations for Images using Attenion) is a multimodal representation learning framework for label-efficient medical image recognition. Our results demonstrate high-performance and label-efficiency for image-text retrieval, classification (finetuning and zeros-shot settings), and segmentation on different medical imaging datasets.
GLoRIA Manuscript
Shih-Cheng Huang (Mars), Liyue Shen, Matthew P. Lungren, Serena Yeung
Stanford University
Proceedings of the IEEE/CVF International Conference on Computer Vision (ICCV), 2021
Start by installing PyTorch 1.7.1 with the right CUDA version, then clone this repository and install the dependencies.
$ conda install pytorch==1.7.1 torchvision==0.8.2 torchaudio==0.7.2 cudatoolkit=10.1 -c pytorch
$ pip install git@github.com:marshuang80/gloria.git
$ conda env create -f environment.ymlMake sure to download the pretrained weights from here and place it in the ./pretrained folder.
import torch
import gloria
# get device
device = "cuda" if torch.cuda.is_available() else "cpu"
# load classifier
num_class = 5 # 5 class classification
freeze = True # freeze encoder and only train linear classifier (less likely to overfit when training data is limited)
model = gloria.load_img_classification_model(num_cls=num_class, freeze_encoder=freeze, device=device)
# load segmentation model (UNet)
seg_model = gloria.load_img_segmentation_model(device=device)import torch
import gloria
import pandas as pd
df = pd.read_csv(gloria.constants.CHEXPERT_5x200)
# load model
device = "cuda" if torch.cuda.is_available() else "cpu"
gloria_model = gloria.load_gloria(device=device)
# generate class prompt
# cls_promts = {
# 'Atelectasis': ['minimal residual atelectasis ', 'mild atelectasis' ...]
# 'Cardiomegaly': ['cardiomegaly unchanged', 'cardiac silhouette enlarged' ...]
# ...
# }
cls_prompts = gloria.generate_chexpert_class_prompts()
# process input images and class prompts
processed_txt = gloria_model.process_class_prompts(cls_prompts, device)
processed_imgs = gloria_model.process_img(df['Path'].tolist(), device)
# zero-shot classification on 1000 images
similarities = gloria.zero_shot_classification(
gloria_model, processed_imgs, processed_txt)
print(similarities)
# Atelectasis Cardiomegaly Consolidation Edema Pleural Effusion
# 0 1.371477 -0.416303 -1.023546 -1.460464 0.145969
# 1 1.550474 0.277534 1.743613 0.187523 1.166638
# .. ... ... ... ... ...This codebase has been developed with python version 3.7, PyTorch version 1.7.1, CUDA 10.2 and pytorch-lightning 1.1.4.
Example configurations for pretraining and downstream classification can be found in the ./configs. All training and testing are done using the run.py script. For more documentation, please run:
python run.py --helpThe preprocessing steps for each dataset can be found in ./gloria/datasets/preprocess_datasets.py
Train the representation learning model with the following command:
python run.py -c ./configs/chexpert_pretrain_config.yaml --train- Please note that the CheXpert radiology reports are still under PHI review for HIPPA compliency, and not publicly availible yet.
Fine-tune the GLoRIA pretrained image model for classification with the following command:
# chexpert
python run.py -c ./configs/chexpert_classification_config.yaml --train --test --train_pct 0.01
# pneumonia
python run.py -c ./configs/pneumonia_classification_config.yaml --train --test --train_pct 0.01The train_pct flag randomly selects a percentage of the dataset to fine-tune the model. This is use to determine the performance of the model under low data regime.
Fine-tune the GLoRIA pretrained image model for segmentation with the following command:
# chexpert
python run.py -c ./configs/pneumothorax_segmentation_config.yaml --train --test --train_pct 0.01@inproceedings{huang2021gloria,
title={GLoRIA: A Multimodal Global-Local Representation Learning Framework for Label-Efficient Medical Image Recognition},
author={Huang, Shih-Cheng and Shen, Liyue and Lungren, Matthew P and Yeung, Serena},
booktitle={Proceedings of the IEEE/CVF International Conference on Computer Vision},
pages={3942--3951},
year={2021}
}
This codebase is adapted from ControlGAN