The disease and gene command utility is a command line developed with Python with two functionalities for retrieving information about diseases and genes.
It was developed with the DISGENET plus API.
- Clone the Repo (or Unzip the .zip file)
git clone https://github.com/mariela-plaza/medbioinformatics-code-test-
Go to the root directory of the project
-
Create Virtual Environment
python -m venv VIRTUAL_ENVIRONMENT_NAME- Activate Virtual Environment
# macOS / linux
source VIRTUAL_ENVIRONMENT_NAME/bin/activate# Windows
VIRTUAL_ENVIRONMENT_NAME\Scripts\activate
- Install Dependencies
pip install -r requirements.txt-
Get an API KEY from https://beta.disgenetplus.com/
-
Enter your API Key in the apiKey.json file in place of "TYPE_YOUR_API_KEY"
{
"apiKey": "TYPE_YOUR_API_KEY"
}From the project directory, open a terminal and use the CLI app as follows:
- For disease information
# Substitute the DISEASE_IDENTIFIER with an UMLS disease code. Examples: C0036341, C0002395, C0013264
python main.py disease DISEASE_IDENTIFIER- For gene variants information
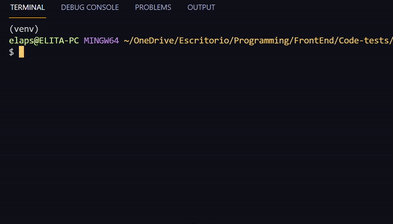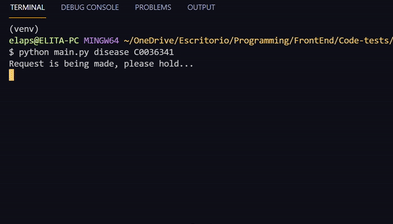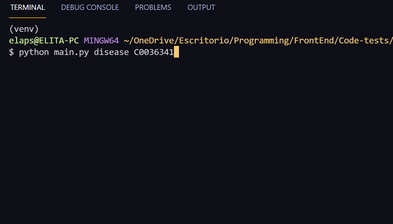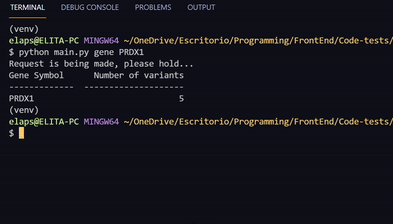
# Substitute the GENE_HGNC_SYMBOL with an HGNC gene symbol. Examples: PRDX1, AGPAT5, LCLAT1
python main.py gene GENE_HGNC_SYMBOL- Mariela Plaza