This is just a sandbox to demonstrate, explore, and vet some ideas I have for developing a better system for curating relevant genomics datasets (mainly annotations) for reproducible research. I am exploring the use of existing tools such as dat and conda for a long term solution. In the interim, this is a place to test ideas and seek feedback.
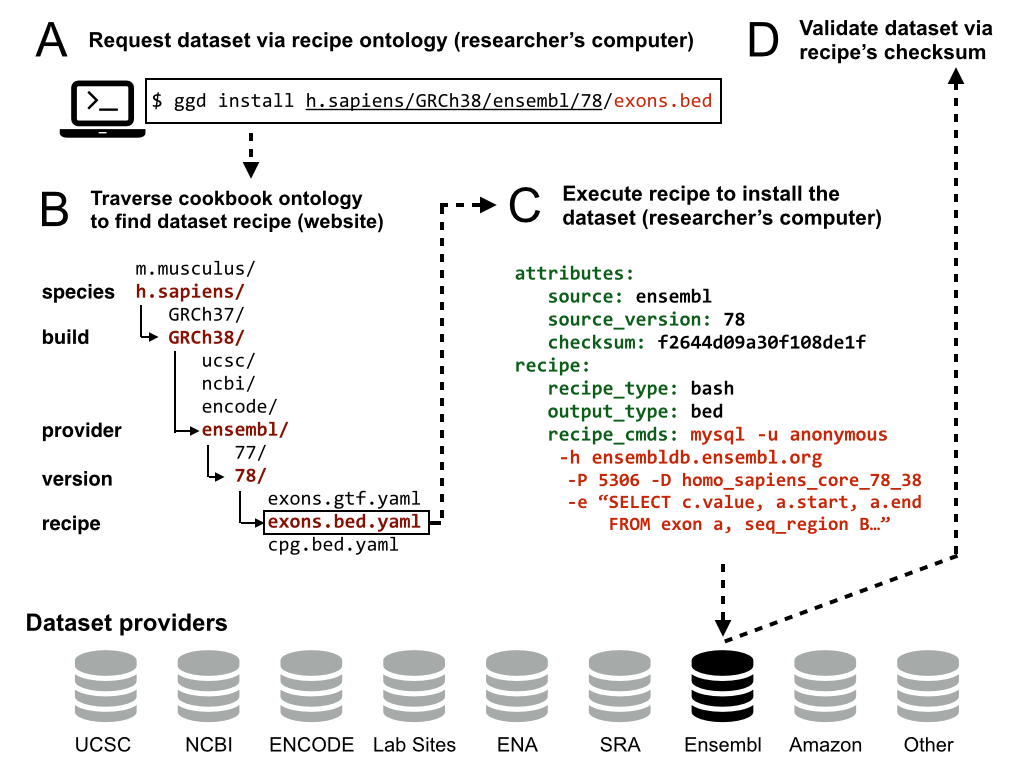
The recipes for datasets are, by default, retrieved from https://github.com/arq5x/ggd-recipes. The idea is to allow this cookbook of recipes to be updated by the research community. Moreover, the GGD software will be able to inspect other "cookbooks" besides the default via the use of the --cookbook option.
The recipes follow an ontology (that is a work in progress). For example, one could use GGD to install CpG islands from UCSC for Human build 38. The command would be:
# Recipe hierarchy: source/species/genomebuild/name
ggd install ucsc/human/b38/cpg
In this case, the recipe would live at:
https://github.com/arq5x/ggd-recipes/ucsc/human/b38/cpg.yaml
Overview of the GGD design (long term vision):
git clone https://github.com/arq5x/ggd
cd ggd
sudo python setup.py install
GGD uses a configuration file ('config.yaml') to control where the datasets that it installs are placed on your file system. By default, this will be $HOME/ggd_data.
Get CpG islands for human build 37 from UCSC
$ ggd install ucsc/human/b37/cpg
searching for recipe: ucsc/human/b37/cpg... ok
validating dataset SHA1 checksum for /Users/arq5x/ggd_data/ucsc.human.b37.cpg... ok ( 08d7fca82507050948b3dfa9c5729895c13b73e6)
installed ucsc/human/b37/cpg
$ ls -1 $HOME/ggd_data
ucsc.human.b37.cpg
If you would the datasets to be placed somewhere else, one can change the path that is used with the setpath tool.
$ ggd setpath --path ~/mydata/
Now get CpG islands for human build 38 from UCSC. Note that the location of the dataset has changed to ~/mydata/
$ ggd install ucsc/human/b38/cpg
searching for recipe: ucsc/human/b38/cpg... ok
validating dataset SHA1 checksum for /Users/arq5x/ggd_data/ucsc.human.b38.cpg... ok ( 733941698f3759f3ade5bc1b6b436ef621200d5c)
installed ucsc/human/b38/cpg
$ ls -1 ~/mydata/
ucsc.human.b38.cpg
Get Clinvar VCF and Tabix index for human build 37 from NCBI
$ ggd install ncbi/human/b37/clinvar
searching for recipe: ncbi/human/b37/clinvar... ok
validating dataset SHA1 checksum for /Users/arq5x/ggd_data/clinvar-latest.vcf.gz... ok ( bd0275c2934b9ea395c5c7e1776ce538bb4e3a1a)
validating dataset SHA1 checksum for /Users/arq5x/ggd_data/clinvar-latest.vcf.gz.tbi... ok ( ec44e825a1f9f3d7f582bb489b66bc82c35ea810)
installed ncbi/human/b37/clinvar
$ ls -1 ~/mydata/
ucsc.human.b38.cpg
clinvar-latest.vcf.gz
clinvar-latest.vcf.gz.tbi
Get a specific genomic region from Clinvar VCF via Tabix for human build 37 from NCBI
$ ggd install ncbi/human/b37/clinvar \
--region 22:20000000-30000000
searching for recipe: ncbi/human/b37/clinvar... ok
installed ncbi/human/b37/clinvar
$ ls -1 ~/mydata/
ucsc.human.b38.cpg
clinvar-latest.region.vcf
clinvar-latest.vcf/gz
clinvar-latest.vcf.gz.tbi
Get ExAC VCF and Tabix index for human build 37 from ExAC website (slower)
$ ggd install misc/human/b37/exac
Try to install a recipe that doesn't exist (nearly everything at this poit)
$ ggd install ucsc/human/b38/dbsnp
searching for recipe: ucsc/human/b38/dbsnp... failed
exiting.
Search for recipes
$ ggd search cpg
Available recipes:
ucsc/human/b37/cpg
ucsc/human/b38/cpg
List all recipes (currently very few)
$ ggd list
misc/human/b37/exac
ncbi/human/b37/clinvar
ucsc/human/b37/alus
ucsc/human/b37/cpg
ucsc/human/b38/cpg
...
Change the config file path defining where data should go:
$ ggd setpath /new/path
Remind yourself of the path where in which the datasets are located:
$ ggd where
We will try to minimize these as much as possible.
- Mysql
- Curl
- pyyaml
- Python 2.7
- Tabix