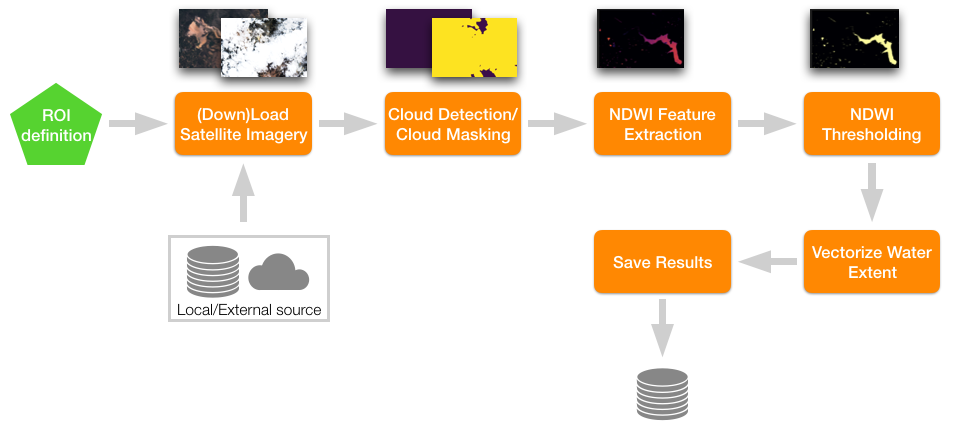
eo-learn makes extraction of valuable information from satellite imagery easy.
The availability of open Earth observation (EO) data through the Copernicus and Landsat programs represents an unprecedented resource for many EO applications, ranging from ocean and land use and land cover monitoring, disaster control, emergency services and humanitarian relief. Given the large amount of high spatial resolution data at high revisit frequency, techniques able to automatically extract complex patterns in such spatio-temporal data are needed.
eo-learn is a collection of open source Python packages that have been developed to seamlessly access and process
spatio-temporal image sequences acquired by any satellite fleet in a timely and automatic manner. eo-learn is
easy to use, it's design modular, and encourages collaboration -- sharing and reusing of specific tasks in a typical
EO-value-extraction workflows, such as cloud masking, image co-registration, feature extraction, classification, etc. Everyone is free
to use any of the available tasks and is encouraged to improve the, develop new ones and share them with the rest of the community.
eo-learn makes extraction of valuable information from satellite imagery as easy as defining a sequence of operations to be performed on satellite imagery. Image below illustrates a processing chain that maps water in satellite imagery by thresholding the Normalised Difference Water Index in user specified region of interest.
eo-learn library acts as a bridge between Earth observation/Remote sensing field and Python ecosystem for data science and machine learning. The library is written in Python and uses NumPy arrays to store and handle remote sensing data. Its aim is to make entry easier for non-experts to the field of remote sensing on one hand and bring the state-of-the-art tools for computer vision, machine learning, and deep learning existing in Python ecosystem to remote sensing experts.
eo-learn is divided into several subpackages according to different functionalities and external package dependencies. Therefore it is not necessary for user to install entire package but only the parts that he needs.
At the moment there are the following subpackages:
eo-learn-core- The main subpackage which implements basic building blocks (EOPatch,EOTaskandEOWorkflow) and commonly used functionalities.eo-learn-coregistration- The subpackage that deals with image co-registraion.eo-learn-features- A collection of utilities for extracting data properties and feature manipulation.eo-learn-geometry- Geometry subpackage used for geometric transformation and conversion between vector and raster data.eo-learn-io- Input/output subpackage that deals with obtaining data from Sentinel Hub services or saving and loading data locally.eo-learn-mask- The subpackage used for masking of data and calculation of cloud masks.eo-learn-ml-tools- Various tools that can be used before or after the machine learning process.
The design of the eo-learn library follows the dataflow programing paradigm and consists of three building blocks:
EOPatch- common data-object for spatio-temporal EO and non-EO data, and their derivatives
EOTask- a single, well-defined action being performed on existing
EOPatch(es) - each
EOTasktakes anEOPatchas an input and returns a modifiedEOPatch
- a single, well-defined action being performed on existing
EOWorkflow- a collection of **
EOTask**s that together represent an EO-value-adding-processing chain
- a collection of **
EOPatch contains multi-temporal remotely sensed data of a single patch (area) of Earth's surface typically
defined by a bounding box in specific coordinate reference system. The EOPatch object can also be used as a placeholder
for all quantities, either derived from the satellite imagery or from some other external source, such as for example
biophysical indices, ground truth reference data, weather data, etc. EOPatch is completely sensor-agnostic, meaning that imagery from different sensors (satellites) or sensor types (optical, synthetic-aperture radar, ...) can be added to an EOPatch.
There's no limitation on the amount of data, or the type of data that can be stored. But typically, all of the information is internally stored in form of NumPy arrays as the following features:
DATA: time- and position-dependent remote sensing data (bands, derived indices, ...) of type double/floatMASK: time- and position-dependent mask (ground truth, cloud/shadow mask, super pixel identifier) of type intDATA_TIMELESS: time-independent and position-dependent remote sensing data (elevation, ...) of type double/floatMASK_TIMELESS: time-independent and position-dependent mask (ground truth, region of interest mask, ...) of type intSCALAR: time-dependent and position-independent remote sensing data (cloud coverage, ...) of type double/floatLABEL: time-dependent and position-independent label (ground truth, ...) of type intSCALAR_TIMELESS: time-independent and position-independent remote sensing data (...) of type double/floatLABEL_TIMELESS: time-independent and position-independent label (ground truth, ...) of type int
The main difference between the different features is in their dimensionality:
DATAandMASKaren_times x height x width x ddimenisional arraysDATA_TIMELESSandMASK_TIMELESSareheight x width x d'dimenisional arraysSCALARandLABELaren_times x d''dimenisional arraysSCALAR_TIMELESSandLABEL_TIMELESSare one dimensional arrays
Above height and width are the numbers of pixels in y and x, d is the number of features (i.e. bands/channels, cloud probability, ...), and n_times
is the number of time-slices (the number of times this patch was recorded by the satellite -- can also be a single image).
Check what kind of features are already in an example EOPatch instance:
eopatch.get_features()Our example EOPatch instance may have for an example:
- a
DATA-feature array namedBANDS-S2-L1Ccontaining 13 Sentinel-2 bands for images of size1013x1029from 88 different dates - a
DATA-feature array namedCLOUD_PROBScontaining cloud probabilities for each frame - a
MASK-feature array namedIS_DATAcontaining valida data masks (indicating which pixels are useful or not) for each frame
In such case the output of the above command would be:
defaultdict(dict,
{<FeatureType.DATA: 'data'>: {'BANDS-S2-L1C': (88, 1013, 1029, 13),
'CLOUD_PROBS': (88, 1013, 1029, 1)},
<FeatureType.MASK: 'mask'>: {'IS_DATA': (88, 1013, 1029)}})Theese arrays can be obtained in two ways:
cloud_probs = eopatch.get_feature(FeatureType.DATA, 'CLOUD_PROBS')or
is_data = eopatch.mask['IS_DATA']Given the information this EOPatch instance has one could calculate for example the cloud mask of each frame and add this derived quantity to the EOPatch instance. The following code-snippet does this:
cloud_probs = eopatch.get_feature(FeatureType.DATA, 'CLOUD_PROBS')
# create cloud mask from cloud probability map by a simple
# threshold (pixel with cloud prob > 0.5 is a cloudy pixel)
cloud_mask = cloud_probs>0.4
# add cloud mask as a MASK-feature
eopatch.add_feature(FeatureType.MASK, 'CLOUD_MASK', cloud_mask)NOTE: Typically the above action would be encapsulated in an EOTask as shown below.
EOPatch can be (de-)serialized (from) to disk. At the moment the EOPatch's features are serialized to disk as NumPy arrays. Subject to change in the future.
**EOTask**s are in a sense the heart of the eo-learn library. They define in what way the available satallite imagery can be manipulated in order to extract valuable information out of it. Typical user will most often just be interested in what kind of tasks are already implemented and ready to be used or how to implement an EOTask, if it doesn't exist yet.
The following **EOTask**s are currently implemented and included into the eo-learn library:
eotask-iosub-package: tasks for downloading, loading, adding features, and serialization of **EOPatch**es- Add Sentinel-2 L1C/L2A and Landsat-8 imagery using Sentinel Hub’s WMS/WCS services
- using predefined layers or/and custom scripts
- Add Digital Elevation Model data
- Add Sen2Cor’s scene classification mask using Sentinel Hub’s WMS/WCS services
- Add raster data (i.e. segmentation masks) from Geopedia
- Burn raster data (i.e. segmentation masks) from vectorised dataset
- Remove features from
EOPatch - Save
EOPatchto disk - Load
EOPatchfrom disk
- Add Sentinel-2 L1C/L2A and Landsat-8 imagery using Sentinel Hub’s WMS/WCS services
eotask-feature-manipulationsub-package: tasks for feature manipulation- Temporal indices of Max/Min of any feature from
EOPatch - Temporal indices of Max/Min slope of any feature from
EOPatch - Spatio-temporal features
- Interpolation of invalid pixels in a time-series
- Temporal indices of Max/Min of any feature from
eotask-masksub-package: task for masking/validating pixels- Cloud masking
- Validating pixels using user's predicate
eotask-coregistrationsub-package: tasks for co-registration of frames in the time series- translational registration using the thunder-registration package
- intensity-based method within opencv
- translational registration using the Elastix library
- point-based registration using opencv-contrib
If a task doesn't exist yet, the user can implement it and easily include it into his/hers workflow. There is very little or almost no overhead in the implementation of a new EOTask as seen from this minimal example:
class FooTask(EOTask):
def __init__(self, foo_param):
self.foo_param = foo_param
def execute(self, eopatch, *, patch_specific_param):
# do what foo does on input eopatch and return it
return eopatchEOTask’s arguments can be static (set when EOTask is initialized; i.e.e foo_param above) or dynamic (set during the execution of the workflow; i.e. patch_specific_param above).
EOWorkflow represents the entire EO processing chain or EO-value-extraction pipeline by chaining or connecting sequence of **EOTask**s. The EOWorkflow takes care that the **EOTask**s are executed in the correct order and with correct parameters. EOWorkflow is executed on a single EOPatch at a time, but the same EOWorkflow can be executed on multiple **EOTask**s.
Under the hood the EOWorkflow builds a directed acyclic graph (in case user tries to build a cyclic graph the EOWorkflow will complain). There's no limitation on the number of nodes (**EOTask**s with inputs) or the graph's topology. The EOWorkflow first names the input tasks that persist over executions, determines the ordering of the tasks, executes the task in that order, and finally returns the results of tasks with no outgoing edge.
The following code snippet shows how to build a very generic workflow to do some basic algebraic operation, such as
# First define simple tasks for each basic operation
class InputNumber(EOTask):
def execute(self, *, input_number):
return input_number
class AddConstant(EOTask):
def __init__(self, constant):
self.constant = constant
def execute(self, number):
return number+self.constant
class Multiply(EOTask):
def execute(self, x, y):
return x * y
class Sum(EOTask):
def execute(self, *numbers):
return sum(numbers)
# Initalize the tasks
in_a = InputNumber()
in_b = InputNumber()
in_c = InputNumber()
add_2 = AddConstant(2)
multi_ab= Multiply()
sum_all = Sum()
# Define the workflow = algebraic operation
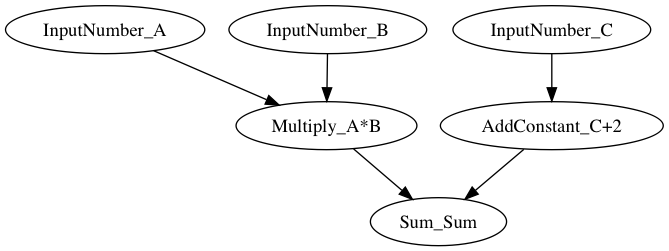
dag = EOWorkflow(dependencies=[
Dependency(transform=in_a, inputs=[]),
Dependency(transform=in_b, inputs=[]),
Dependency(transform=in_c, inputs=[]),
Dependency(transform=multi_ab, inputs=[in_a, in_b]),
Dependency(transform=add_2, inputs=[in_c]),
Dependency(transform=sum_all, inputs=[multi_ab, add_2])],
task2id={in_a:'A', in_b:'B', in_c:'C',
multi_ab:'A*B', add_2:'C+2', sum_all:'Sum'}
)
# Before executing it, let's look at the execution order
dag.orderwhich is
['A', 'C', 'C+2', 'B', 'A*B', 'A*B + C + 2']Now, let's execute it for the input
dag.execute({'A':{'input_number':2},
'B':{'input_number':3},
'C':{'input_number':5}})The result is
{'Sum': 13}Execute it again on a different input
dag.execute({'A':{'input_number':5},
'B':{'input_number':3},
'C':{'input_number':2}})and the results is
{'Sum': 19}The graph can also be visualized using graphviz:
Users should eventually interact only with the EOWorkflow -- define it and execute it over region of interest -- and not with implementation of **EOTask**s.
Each of the subpackages can be installed separately
pip install git+https://github.com/sentinel-hub/eo-learn#subdirectory=core --upgrade
pip install git+https://github.com/sentinel-hub/eo-learn#subdirectory=coregistration --upgrade
pip install git+https://github.com/sentinel-hub/eo-learn#subdirectory=features --upgrade
pip install git+https://github.com/sentinel-hub/eo-learn#subdirectory=geometry --upgrade
pip install git+https://github.com/sentinel-hub/eo-learn#subdirectory=io --upgrade
pip install git+https://github.com/sentinel-hub/eo-learn#subdirectory=mask --upgrade
pip install git+https://github.com/sentinel-hub/eo-learn#subdirectory=ml_tools --upgradeTo install all packages at once you can download the repository and call
python install_all.pyFor more information on the package content, visit readthedocs.
See LICENSE.