Activation Function Player with PyTorch.
This repository is the implementation of paper AReLU: Attention-based-Rectified-Linear-Unit.
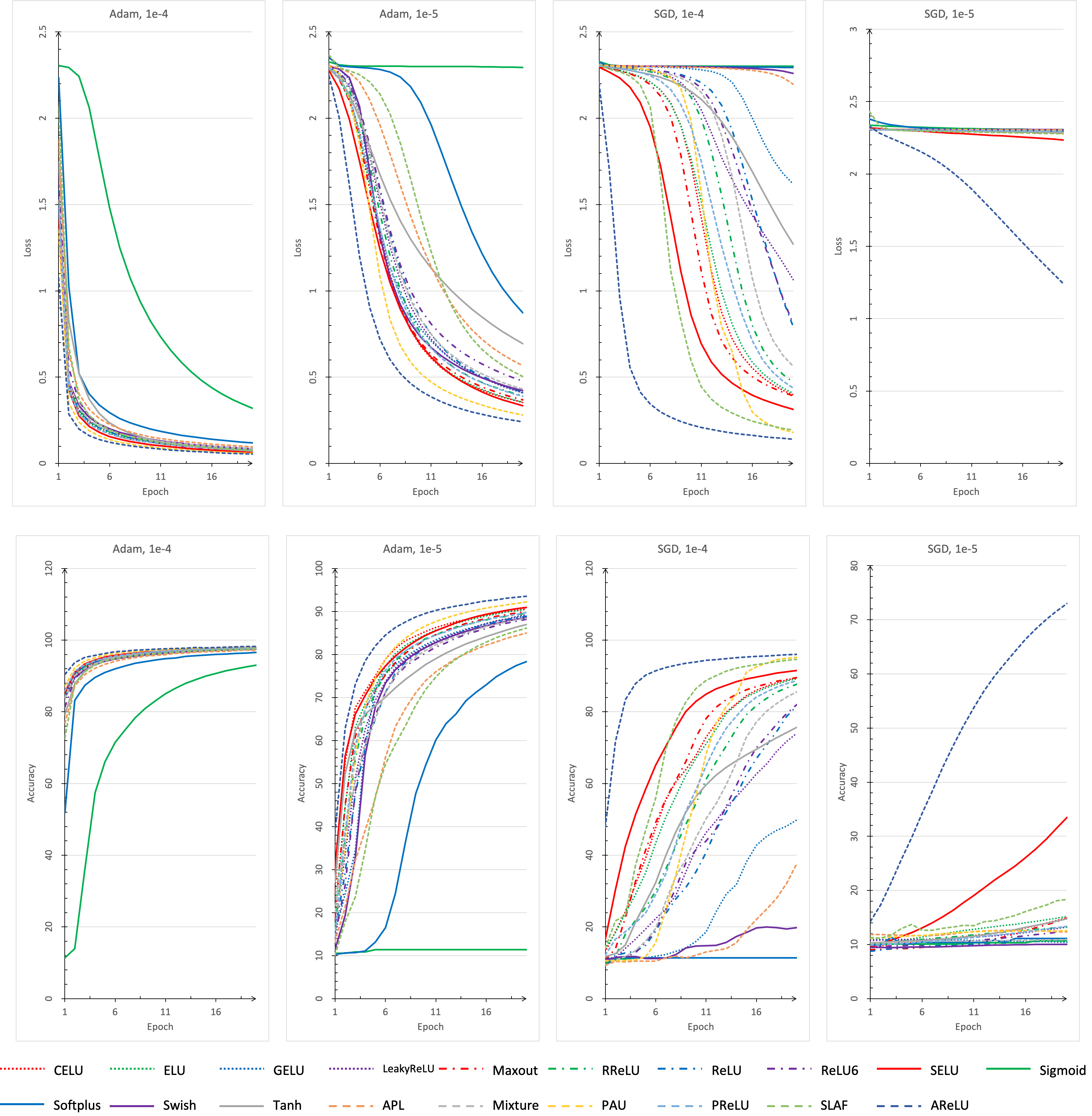
While developing, we found that this repo is quite convenient for people doing experiments with different activation functions, datasets, learning ratings, optimizers and network structures. It is easy for us to add new activation functions and network structures into program. What's more, based on visdom and ploty, a nice visualization of training procedure and testing accuracy has been provided.
This project is friendly to newcomers of PyTorch.
conda create -n AFP python=3.7 -y
conda activate AFP
pip install -r requirements.txtNOTE: PAU is only CUDA supported. You have to compile it first:
pip install airspeed==0.5.14
cd activations/pau/cuda
python setup.py installThe code of PAU is directly token from PAU, if you occur any problems while compiling, please refer to the original repository.
If you just want to have a quick start, and do not want to compile with PAU, just comment out the following lines in activations/__init__.py:
try:
from .pau.utils import PAU
__class_dict__["PAU"] = PAU
except Exception:
raise NotImplementedError("")We use visdom to visualize training process. Before training, please setup visdom server:
python -m visdom.server &Now, you can click here to check your training loss and testing accuracy while runtime.
NOTE: Don't worry about training data. The program will download dataset while runtime and save it under args.data_root
If you want to have a quick start with default parameters, just run:
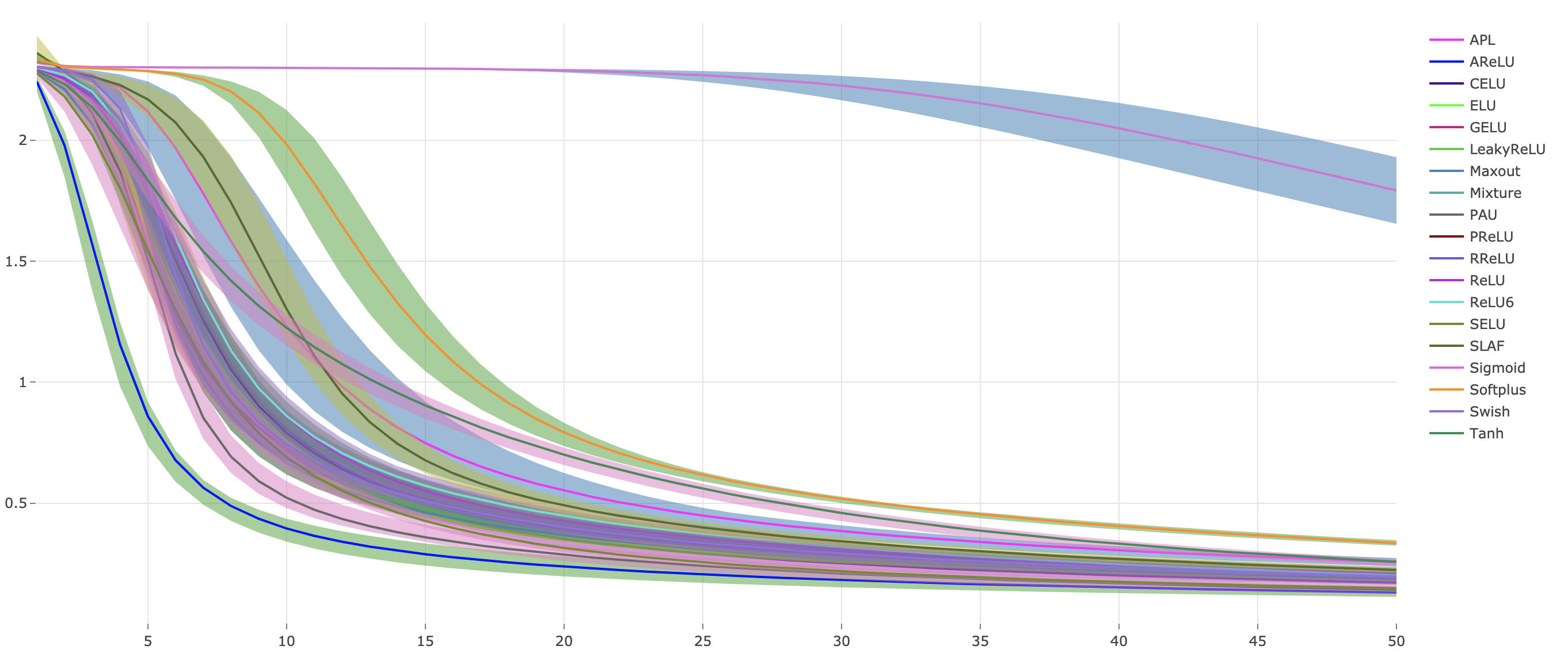
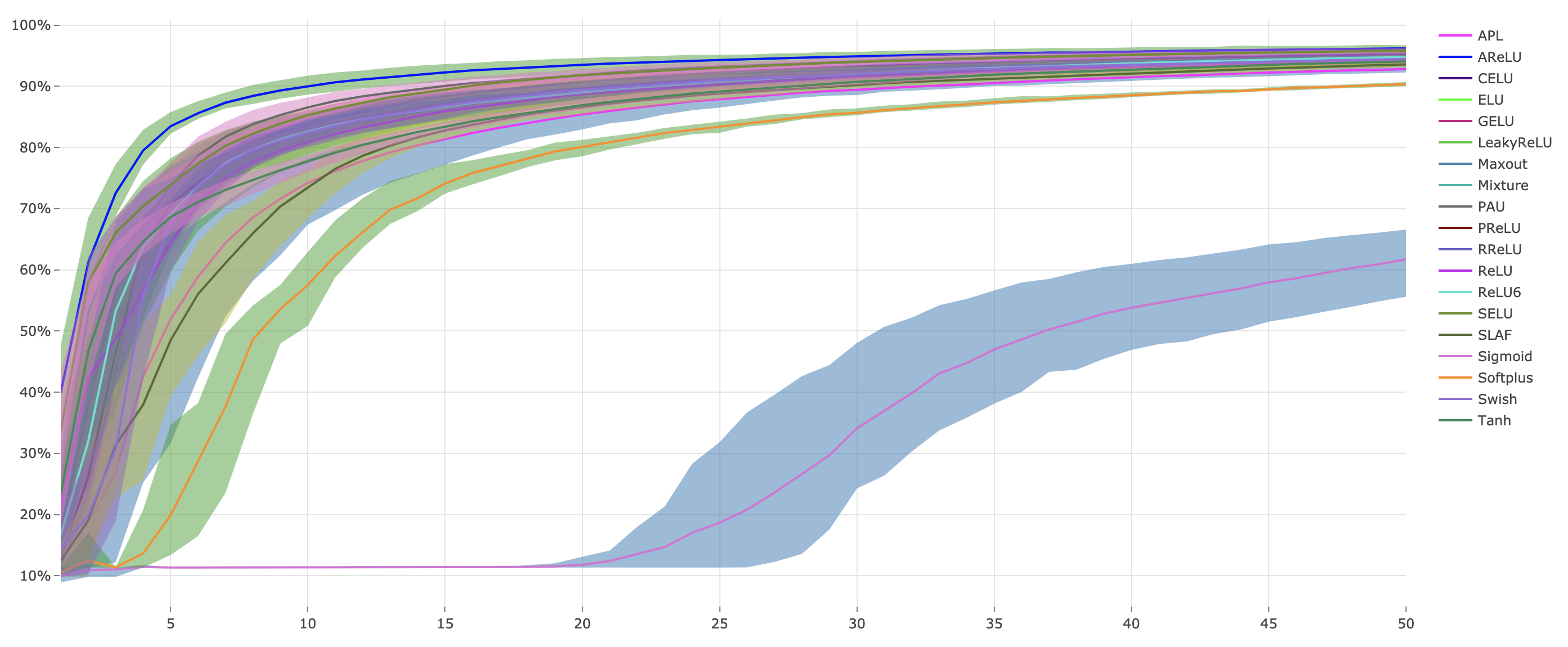
python main.py --cudaWe plot the Continuous Error Bars with ploty and save it as a html file under results folder.
A json file which records same static data is also generated and saved under results.
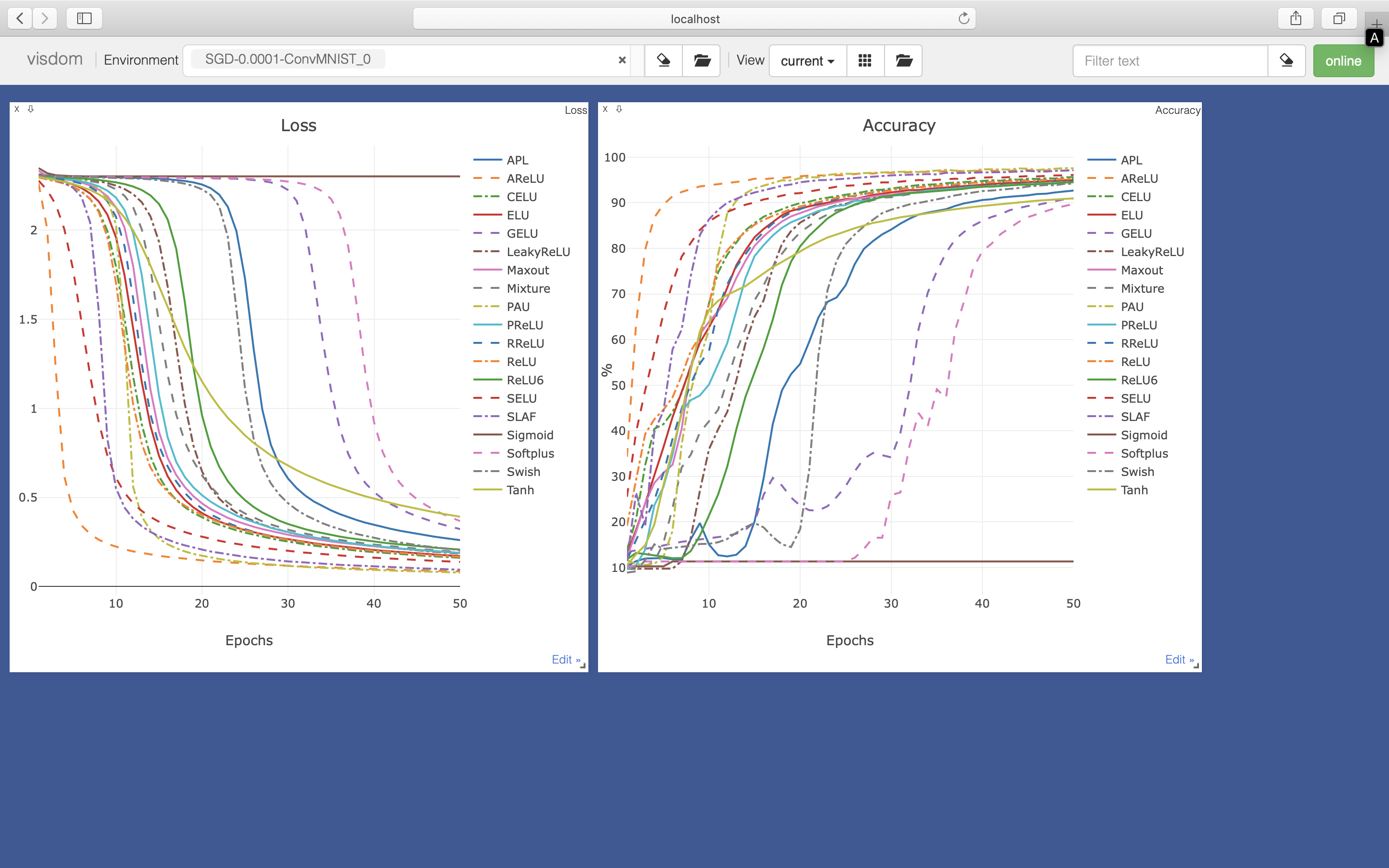
Training loss (visualzie on visdom: http://localhost:8097/):
Testing accuracy (visualize on visdom: http://localhost:8097/):
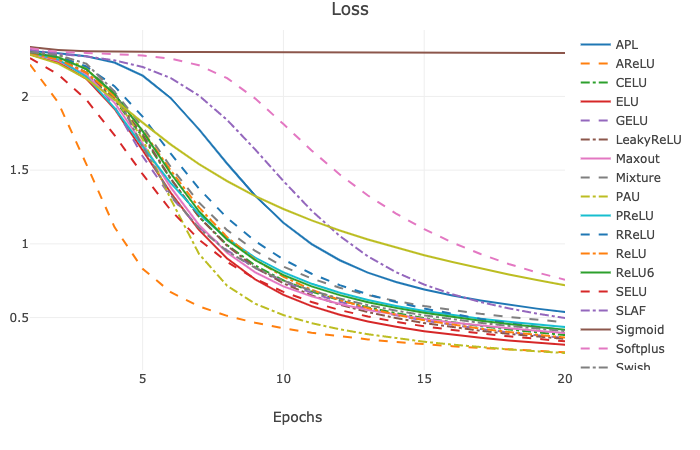
Continuous Error Bars of training loss with five runs (saved under results as html file):
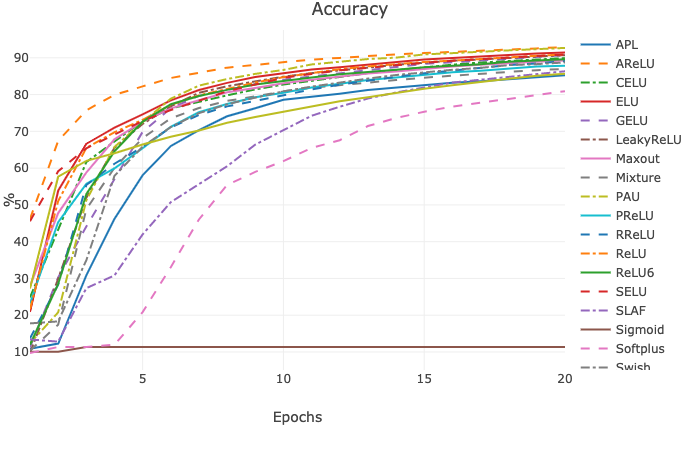
Continuous Error Bars of testing accuracy with five runs (saved under results as html file):
python main.py -h
usage: main.py [-h] [--batch_size BATCH_SIZE] [--lr LR] [--epochs EPOCHS]
[--times TIMES] [--data_root DATA_ROOT]
[--dataset {MNIST,SVHN}] [--num_workers NUM_WORKERS]
[--net {BaseModel,ConvMNIST,LinearMNIST}] [--resume RESUME]
[--af {APL,AReLU,GELU,Maxout,Mixture,SLAF,Swish,ReLU,ReLU6,Sigmoid,LeakyReLU,ELU,PReLU,SELU,Tanh,RReLU,CELU,Softplus,PAU,all}]
[--optim {SGD,Adam}] [--cuda]
[--exname {AFS,TransferLearningPretrain,TransferLearningFinetune}]
Activation Player with PyTorch.
optional arguments:
-h, --help show this help message and exit
--batch_size BATCH_SIZE
batch size for training
--lr LR learning rate
--epochs EPOCHS training epochs
--times TIMES repeat runing times
--data_root DATA_ROOT
the path to dataset
--dataset {MNIST,SVHN}
the dataset to play with.
--num_workers NUM_WORKERS
number of workers to load data
--net {BaseModel,ConvMNIST,LinearMNIST}
network architecture for experiments. you can add new
models in ./models.
--resume RESUME pretrained path to resume
--af {APL,AReLU,GELU,Maxout,Mixture,SLAF,Swish,ReLU,ReLU6,Sigmoid,LeakyReLU,ELU,PReLU,SELU,Tanh,RReLU,CELU,Softplus,PAU,all}
the activation function used in experiments. you can
specify an activation function by name, or try with
all activation functions by `all`
--optim {SGD,Adam} optimizer used in training.
--cuda with cuda training. this would be much faster.
--exname {AFS,TransferLearningPretrain,TransferLearningFinetune}
experiment name of visdom.We provide a script for doing a full training with all activation functions, learning rates, optimizers and network structures.
Just run:
./train.shNOTE: This step is time consuming.
-
write a python script file under
activations, such as new_activation_functions.py, where contains the implementation of new activation function. -
import new activation functions in activations/__init__.py, like:
from .new_activation_functions import NewActivationFunctions
-
Enjoy it!
-
Write a python script file under
models, such as new_network_structure.py, where contains the definition of new network structure. New defined network structure should be a subclass of BaseModel, which defined inmodels/models.py. Such as:from models import BaseModel import torch import torch.nn as nn import torch.nn.functional as F class LinearMNIST(BaseModel): def __init__(self, activation: nn.Module): super().__init__(activation) self.linear1 = nn.Sequential( nn.Linear(28 * 28, 512), activation(), ) self.linear2 = nn.Sequential( nn.Linear(512, 10), nn.LogSoftmax(dim=-1) ) def forward(self, x): x = x.view(-1, 28 * 28) x = self.linear1(x) x = self.linear2(x) return x
-
Import new network structure in models/__init__/py, like:
from .conv import ConvMNIST
-
Enjoy it!
You can modify main.py to try with more datasets and optimizers.
You can refer to CIFAR10 and CIFAR100 for more experiments with popular network structures.
After downloading the repo, you just copy activations folder into repo, and modify some code.
You can refer to Detectron2 for more experiments on segmentation. And refer to UNet-Brain for a simple test with UNet on brain segmentation.
We provide a simple script to play with transfer learning between MNIST and SVHN.
./transfer_learning.shWe provide a lightly python script that can collect the json file data which generated under result folder to readable latex code.
python json_to_latex.py -h
usage: json_to_latex.py [-h] [--exname EXNAME] [--data {best,mean,std}]
[--epoch {first epoch,best}] [--output OUTPUT]
Json to LaTex (Lightly)
optional arguments:
-h, --help show this help message and exit
--exname EXNAME exname to generate json
--data {best,mean,std}
best: best accuracy, mean: mean accuracy, std: std of
acc
--epoch {first epoch,best}
which epoch to load.
--output OUTPUT output filename