A Framework for Comparing and Selecting from Several Segmentation Methods in Absence of Ground Truth labels
Find the bioRxiv pre-print here!
And our poster from the HTAN Summer Meeting 2022 here!
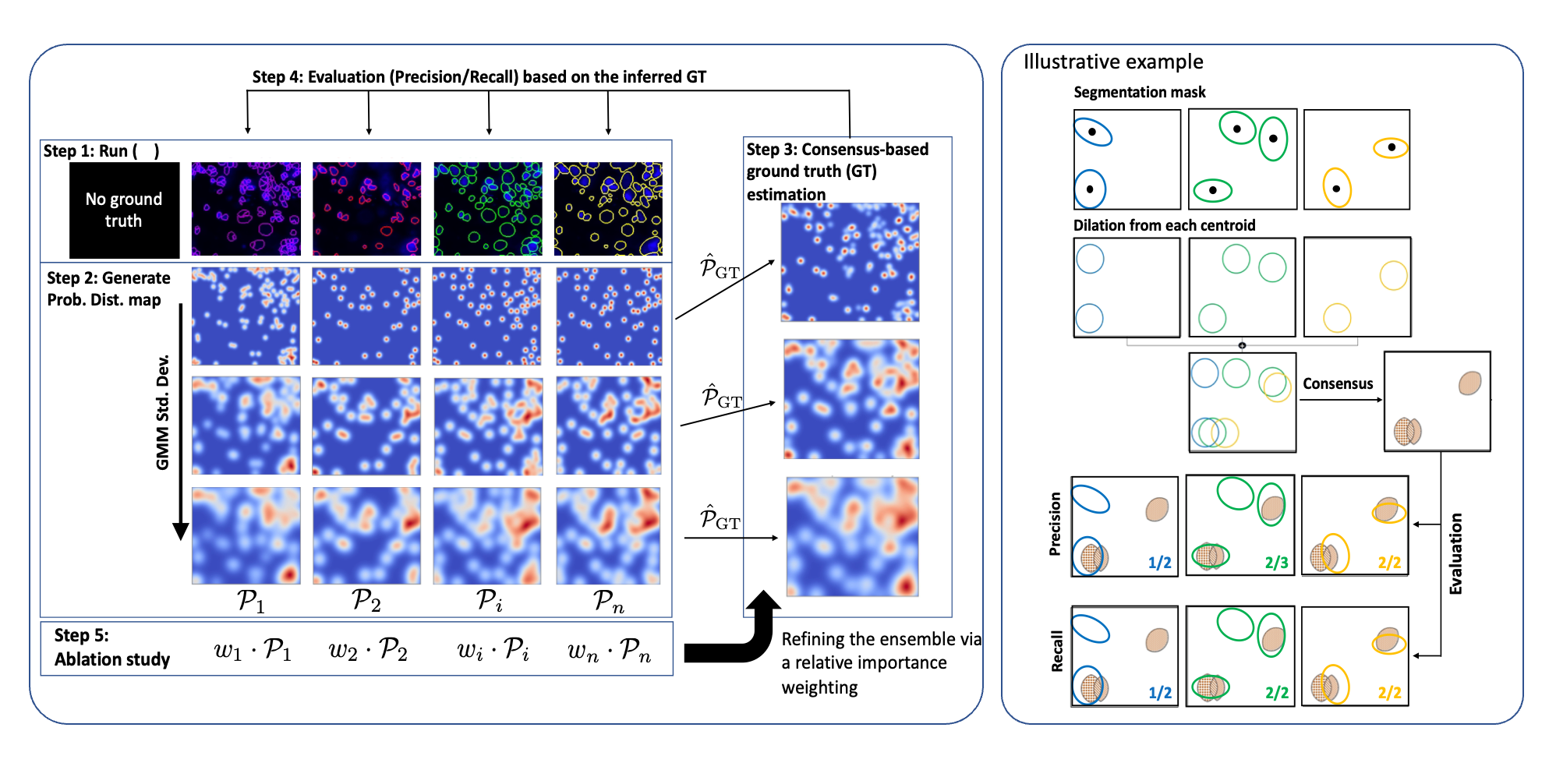
Our method derives a consensus-based ground truth segmentation by combining and ablating the results from multiple candidate segmentations. The process is probabilistic, interpreting each segmented object as a spatial probability distribution of a true segmentation. Probability maps are combined and and resulting "ground-truth" map is derived, which is then used to iteratively weight the input segmentations and improve the overall agreement across methods as represented by the ground-truth result. The below figure illustrates this process in detail:
The Segmentation Evaluation Methods Breakdown.ipynb notebook gives a step-by-step breakdown of the process used in the accompanying paper.