Lukas Muenter 27 7 2021
NOTE: Currently, this package does only accept AGI-codes (A. thaliana). This will change, however.
This package provides a client for GO-Term enrichment via the API of
PANTHER. It takes a vector of gene IDs,
sends it to PANTHER, and reformats the response into a handy
dataframe. This dataframe also includes gene IDs, which are
associated to the GO-Term in question.
# install from github
devtools::install_github("lmuenter/oracl")
In this example, we’d like to identify overrepresented GO-Terms for an
example dataset provided with the package. Note, that we specify the
Biological Process ontology by setting ont = bp in
oracl::oraclient(). Other options are of course ont = mf (Molecular
Function) and ont = cc (Cellular Component).
# load package
library(oracl)
# Get a set of AGI-codes.
gs <- oracl:::GS01
# Get a background geneset (optional)
bg <- oracl:::background
# conduct GO-Term ORA via PANTHER
bp.df <- oraclient(gs, bg = bg, ont = "bp", fdr.thresh = 0.05)## Joining, by = "GO_ID"
# Load Packages
library(ggplot2)
# Make a plot
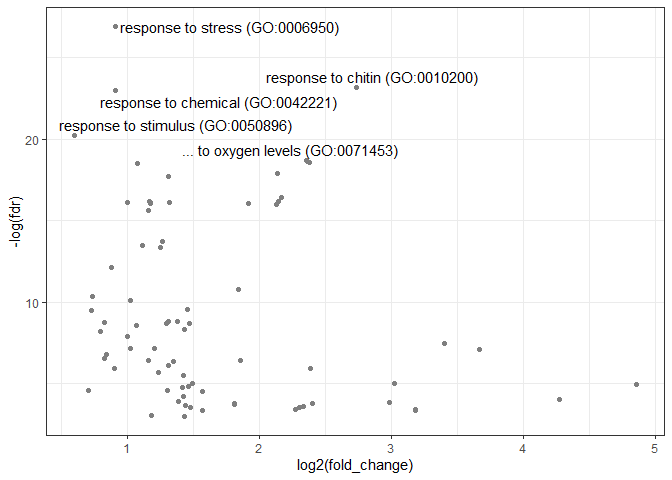
volcano.p = volcanoracl(bp.df)## Adding missing grouping variables: `grouping`
## Joining, by = c("label", "grouping")
# The plot `volcano.p` is a ggplot-object.
# We can change its attributes!
volcano.p + scale_colour_gradientn(colours = "steelblue")When several genesets should be inferred, it may be handy to combine overrepresented terms in one dataframe. This is especially useful for plotting.
# obtain a list of genesets
gs.ls <- list(
oracl:::GS01,
oracl:::GS02,
oracl:::GS03
)
# get background geneset
bg <- oracl:::background
# set names of list elements (vital for later)
names(gs.ls) <- c("GS01", "GS02", "GS03")
# get overrepresented GO-terms
bp.ls = lapply(gs.ls, oraclient,
bg = bg,
ont = "bp",
fdr.thresh = 0.05
)## Joining, by = "GO_ID"
## Joining, by = "GO_ID"
## Joining, by = "GO_ID"
# get ONE dataframe (ID-column `grouping` specifies the geneset)
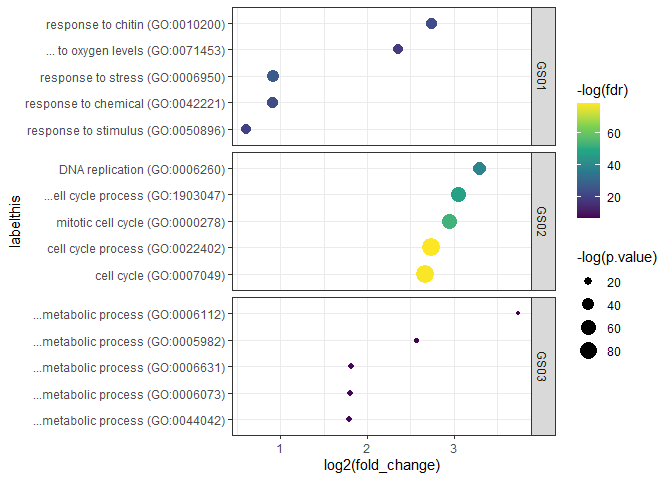
bp.ls.df <- oracl_list_to_df(bp.ls)We can now plot overrepresented GO-Terms using group information in the
column bp.df$grouping. Here, we want to facet the plot according to
the grouping variable (stored in bp.df$grouping). We also specify the
desired number of columns, the position of the facet label, and whether
or not we only include labels found in each dataset (change these things
according to your data!):
oraclot(bp.ls.df, top_n = 5) +
facet_wrap(grouping ~ ., ncol = 1, strip.position = "right", scales = "free_y") +
scale_color_viridis_c()## Adding missing grouping variables: `grouping`
## Joining, by = c("label", "grouping")
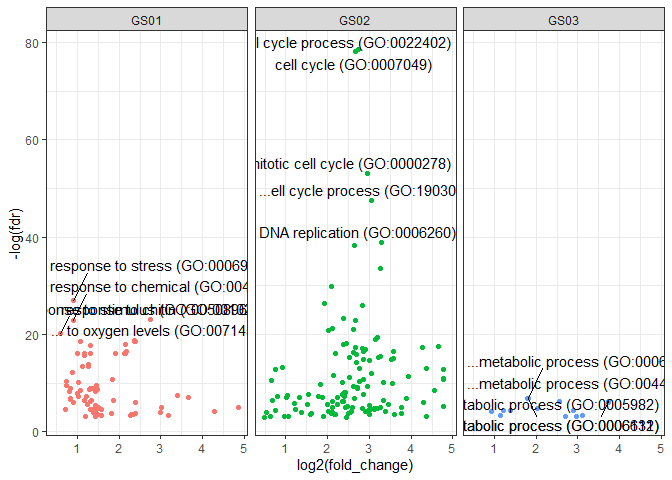
We can also make a facetted volcano plot:
volcanoracl(bp.ls.df, top_n = 5) +
facet_wrap(grouping ~ ., nrow = 1)## Adding missing grouping variables: `grouping`
## Joining, by = c("label", "grouping")
-
Gene IDs and Organism. Currently, only Arabidopsis thaliana (L.) Heynh. can be investigated.
-
Cognate genes. in order to save resources, the API of
PANTHERdoes not report gene sets back (personal communication). Gene IDs reported by{oracl}are therefore only approximations. In essence, the underlying geneset is semantically compared to a gene-to-GO-term-dataset for every enriched GO-Term. These datasets are included in{oracl}(seeoracl/data/goterms). Datasets have been generated by conducting ORA using the PANTHER website with all available AGI codes. To obtain necessary datasets, all results (without Bonferroni Correction) were exported to .json, parsed, and reformated.
-
Functions for automated plotting.
-
Make other organisms available.
-
Implement redundancy removal using
{rrvgo} -
Automate gene-symbol mapping using
{org.At.tair.db}