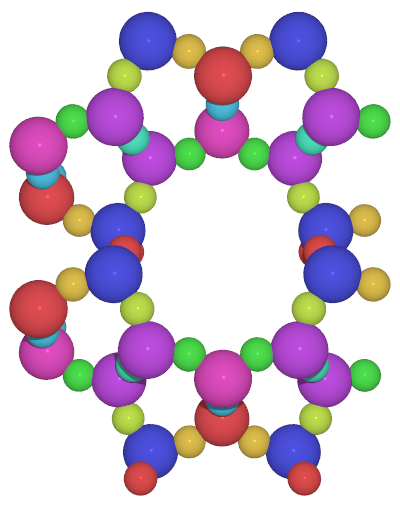
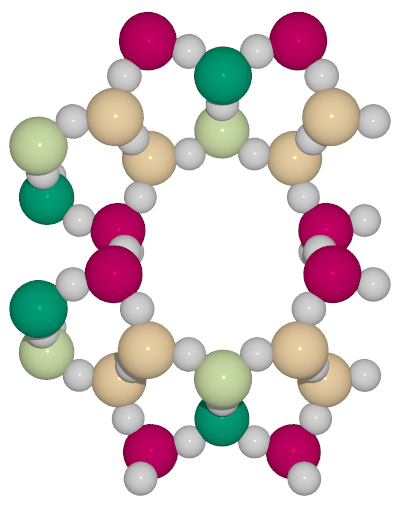
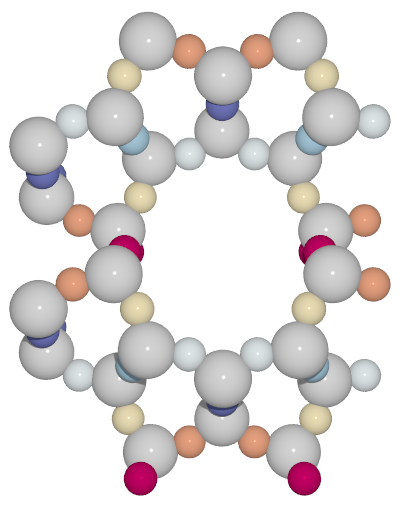
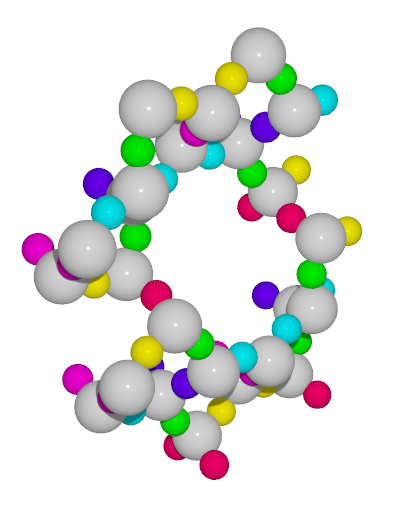
A small utility python script to generate high-quality images from cif files with symmetry unique atoms colored with different colors
First make sure you have the following dependencies installed:
After downloading and unpacking or cloning the repository run the standard
setuptools command
python setup.py install [--user]The scipt takes the following optional arguments that can be display with the standard -h flag:
colorcif.py -h- color all symmetry unique atoms
colorcif.py TON.cif -o ton_default.pov- color only T-atoms using
RdYlGncolormap from matplotlib
colorcif.py -T -c RdYlGn -o ton_RdYlGn_T.pov TON.cif- color only oxygen atoms using
RdYlBucolormap from matplotlib
colorcif.py -O -c RdYlBu -o ton_RdYlBu_O.pov TON.cif- color only oxygen atoms using
gist_rainbowcolormap from matplotlib and rotate the unit cell 30 degrees with respect to each (x, y, z) axes
colorcif.py -O -c gist_rainbow -o ton_gist_rainbow_O.pov -x 30 -y 30 -z 30 TON.cif-
color only oxygen atoms using
gist_rainbowcolormap from matplotlib and rotate the unit cell 30 degrees with respect to each (x, y, z) axes and theglasstexturecolorcif.py -O -c gist_rainbow -o ton_gist_rainbow_O.pov -x 30 -y 30 -z 30 -t glass TON.cif
The script is based on the example from the ase documentation.