The goal of GSEAdv is to provide methods to work with gene sets collections.
This package is abandonded as a new package is being developed to work with gene sets under Bioconductor. My own efforts are now in BaseSet
GSEAdv is based on the relationship between genes and gene sets under this schema: 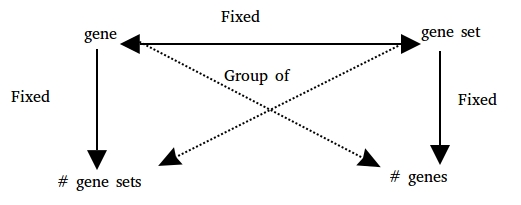
And provides methods to understand the relationships between each property of the schema and as a whole.
It is an R package you will be able to install it from the Bioconductor project with:
## install.packages("BiocManager")
BiocManager::install("GSEAdv")You can install this version of GSEAdv with:
## install.packages("devtools")
devtools::install_github("llrs/GSEAdv")It is simple, load the package and learn from your data!
# Load some data
library("GSEAdv")
fl <- system.file("extdata", "Broad.xml", package = "GSEABase")
gss <- getBroadSets(fl)
gss
## GeneSetCollection
## names: chr5q23, chr16q24 (2 total)
## unique identifiers: ZNF474, CCDC100, ..., TRAPPC2L (215 total)
## types in collection:
## geneIdType: SymbolIdentifier (1 total)
## collectionType: BroadCollection (1 total)
summary(gss)
## Genes: 215
## Gene in more pathways: 1 pathways
## h-index: 0 genes with at least 0 pathways.
## Pathways: 2
## Biggest pathway: 129 genes
## h-index: 1 pathways with at least 1 genes.
## All genes in a single gene set.Which tells us that each gene in the GeneSetCollection is only on one gene set.
We can try with a bigger dataset, one derived from human genes pathways in KEGG:
summary(genesKegg)
## Genes: 5869
## Gene in more pathways: 51 pathways
## h-index: 0 genes with at least 0 pathways.
## Pathways: 228
## Biggest pathway: 1130 genes
## h-index: 15 pathways with at least 15 genes.
## IC(genesPerPathway): 6.65 ( 0.96 of the maximum)
## IC(pathwaysPerGene): 2.47 ( 0.48 of the maximum)Knowing that it has so much pathways and genes we can learn how do they relate. The number of genes per pathway in the collection is:
gpp <- genesPerPathway(genesKegg)
plot(table(gpp))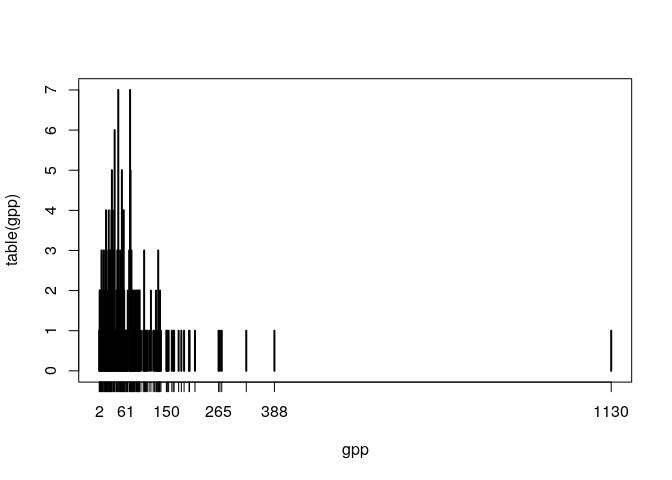 We can see that most gene sets have low number of genes but one has 1130 genes in a single gene set (It is the gene set 01100). The genes might be associated too with many gene sets, it is so extreme? Let's see:
We can see that most gene sets have low number of genes but one has 1130 genes in a single gene set (It is the gene set 01100). The genes might be associated too with many gene sets, it is so extreme? Let's see:
ppg <- pathwaysPerGene(genesKegg)
plot(table(ppg))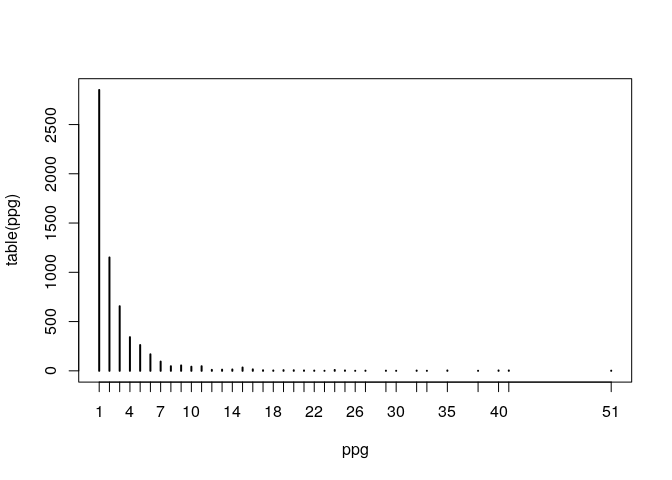 Not so extreme, one gene (5594) appears in 51 gene sets.
Not so extreme, one gene (5594) appears in 51 gene sets.
To see which gene sets are included in other gene sets we can use nested:
nested(genesKegg)[1:10, 80:90]
## 00970 00980 00982 00983 01040 01100 02010 03008 03010 03013 03015
## 00010 0 0 0 0 0 0 0 0 0 0 0
## 00020 0 0 0 0 0 1 0 0 0 0 0
## 00030 0 0 0 0 0 0 0 0 0 0 0
## 00040 0 0 0 0 0 0 0 0 0 0 0
## 00051 0 0 0 0 0 0 0 0 0 0 0
## 00052 0 0 0 0 0 0 0 0 0 0 0
## 00053 0 0 0 0 0 0 0 0 0 0 0
## 00061 0 0 0 0 0 1 0 0 0 0 0
## 00062 0 0 0 0 0 1 0 0 0 0 0
## 00071 0 0 0 0 0 0 0 0 0 0 0As expected the pathway with more than 1100 genes has other pathways inside it.
You can see the vignettes for more examples.
It is intended for bioinformaticians, both people interested in comparing databases and people developing analysis using the information provided by GSEAdv.
The goal of this project is to be able to understand the gene sets collections available.
- Measure properties of the genes and pathways given a
GeneSetCollectionThe number of genes by pathway, the probability of having x genes in more than y pathways... - Compare pathway database: By comparing the differences between them.
- Select the gene set collection of interest: By testing their properties.
- Create
GeneSetCollections with certain properties Create aGeneSetCollectionwere the collections follow certain distributions.
Please read how to contribute for details on the code of conduct, and the process for submitting pull requests.
You can also look at the tests and add more tests to increase the quality of the package.
The ideas of this package were developed after a colleague asked a question in a poster presentation of my other package BioCor. To know the whole history you can read this blogpost.
