Visualization of data is required for most scientific research. Based on visual graphics, we can better display the major features of scientific data. The clinical medical students and junior researchers frequently need to spend dozens of days or even months to learn visualization tools, such as SPSS, Origin, and Graphpad. If you want to specialize in data analysis and modeling, you need to learn one or more programming languages (such as MATLAB, R, and Python). To reach the level where you can freely explore data, you need to spend more time on in-depth learning and advanced.
In recent years, with the development of various cloud computing platforms (such as Galaxy and DNAnexus in the biomedical field) and IT hardware and software (such as distributed computing, container technology, package manager, workflow language, etc.), junior researchers can easily obtain the upstream result. Moreover, when the upstream analysis process of conventional omics data tends to be stable and perfect, the customization and variability of the upstream analysis process have been greatly reduced. The visualization and personalized in-depth interpretation in the downstream process of data analysis have become the biggest challenge facing current users:
- Most of the visualization software or methods developed by the open source user community have not been well integrated under a unified user interface;
- There is no active collaborative community for the visualization of scientific research data in China, and the "** Draw Group" has become one of the few choices for primary scientific research users;
- There is a lack of core data visualization software and platforms similar to Graphpad and MATLAB in China. After being banned by the United States, they can only spend additional costs to migrate the process or start over;
- There are several disadvantages of existing platforms and tools: the user interface is not beautiful enough; lack of the Chinese and English support; it is still difficult to get started; the file Manager of some platforms is not convenient; users cannot actively participate in platform development;
- Data visualization tools are still relatively scarce.
Hiplot is one of the community-driven bioinformatics data visualization platforms. This project started in October 2019 and accelerate in 2020 after COVID-19 breakout. It consists of a 6-person core team covering the IT, bioinformatics students, statistical analysts, and clinicians. The interdisciplinary cooperation may help us for better development of this application. The major development goals of Hiplot:
- Provide the simple and easy-to-use Web and RESTful APIs for the bioinformatics data visualization tools that are no graphical interface support yet.
- Features cover the basic statistics and visualization, predictable clinical models, data analysis of macrobiotics, and the bioinformatics applications of deep learning methods
- Building a collaborative bioinformatics community for the data visualization
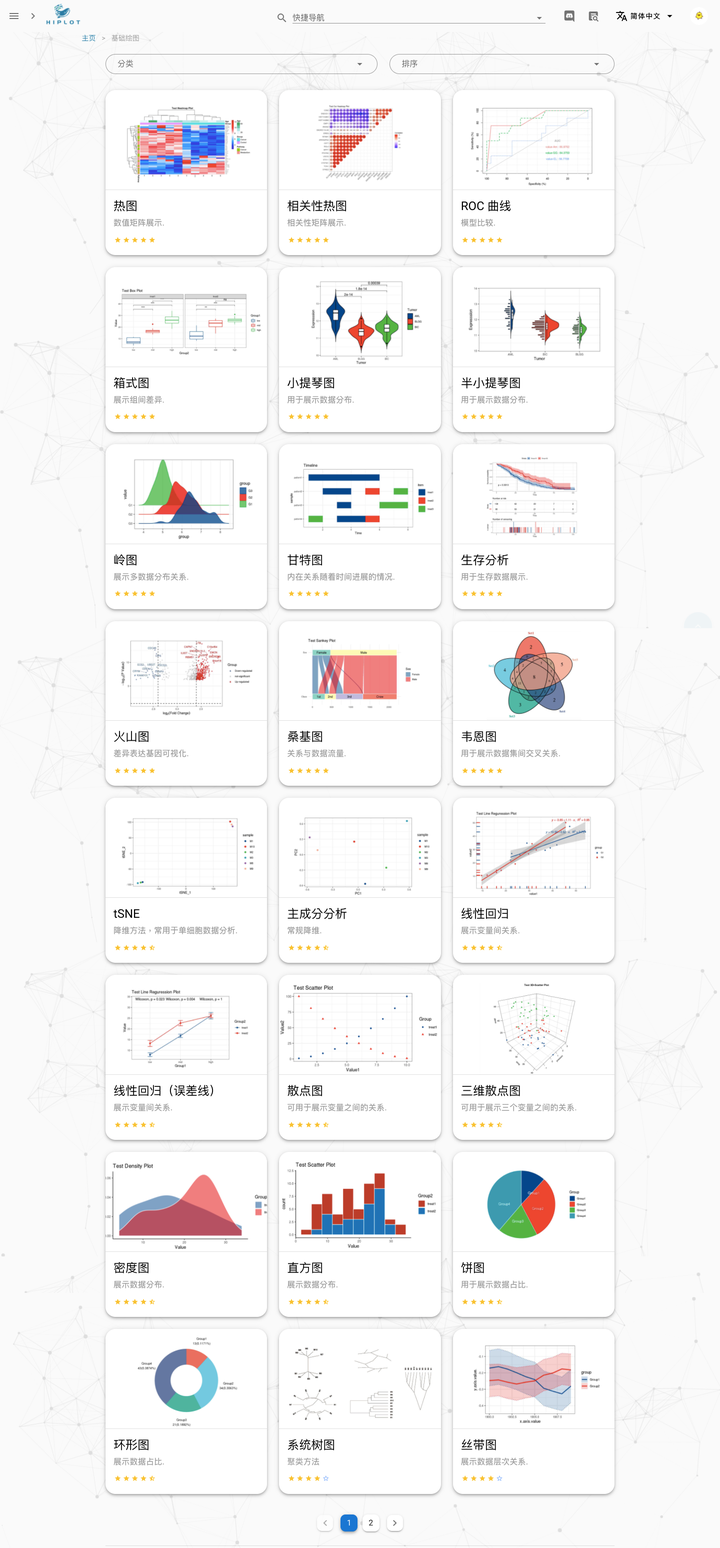
Untill now, we have been developed more than 40 visualization plugins on the Hiplot platform and are still increasing:
User Interface Examples:
Login
Signup
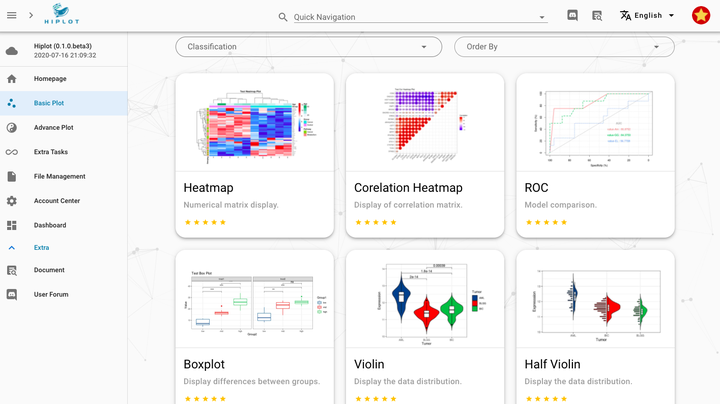
Overview of basic plots
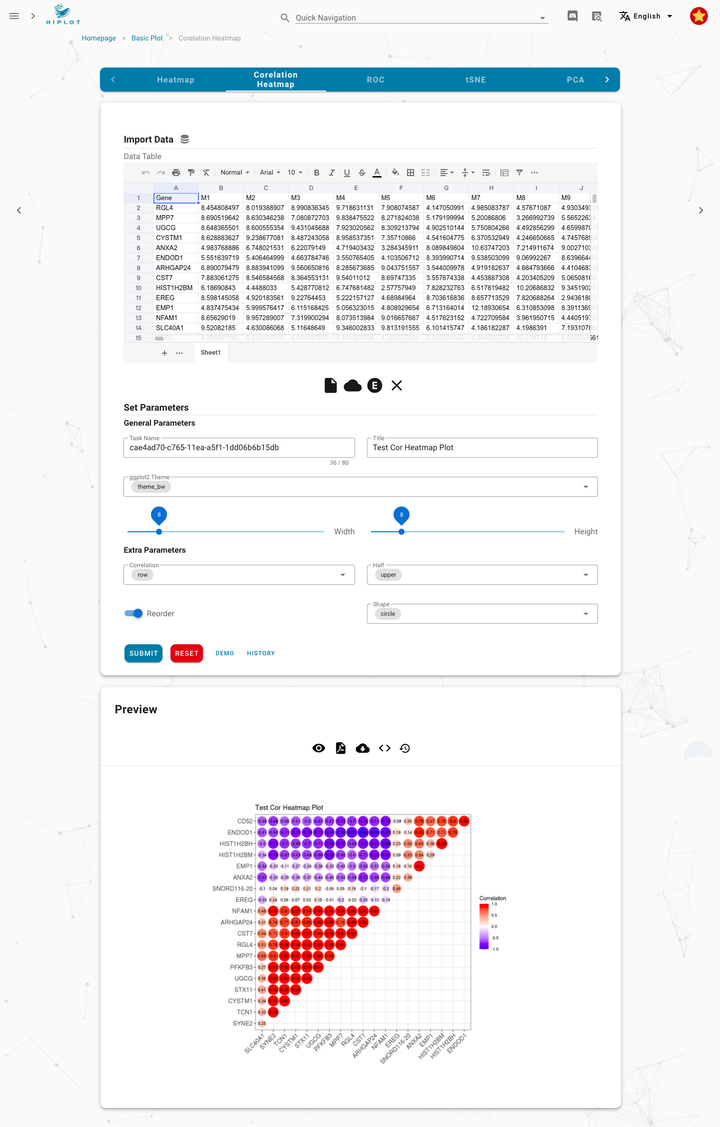
Demo | Correlation heatmap
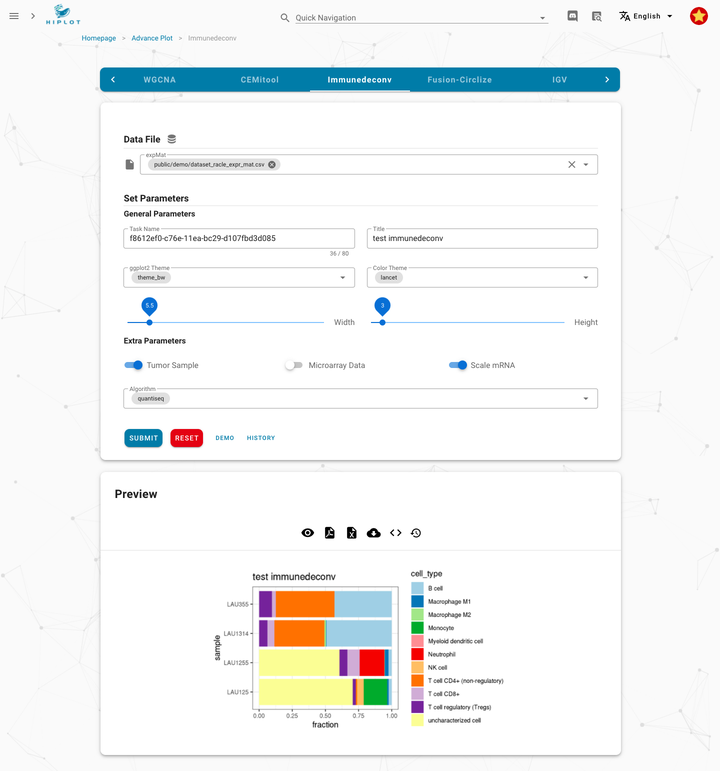
Demo | Immune Infiltration Analysis
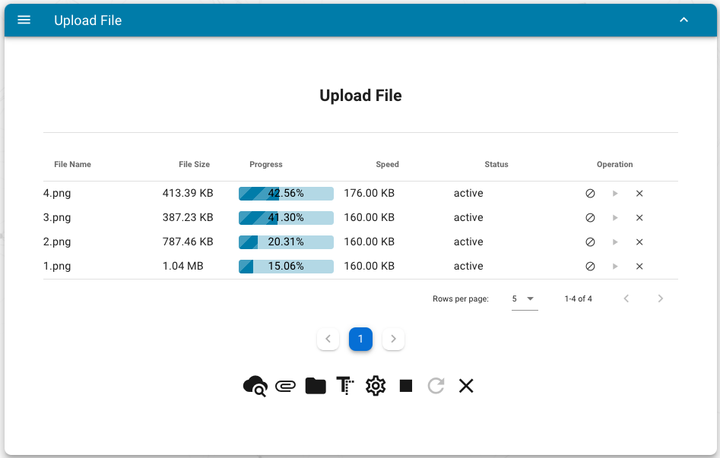
Upload files
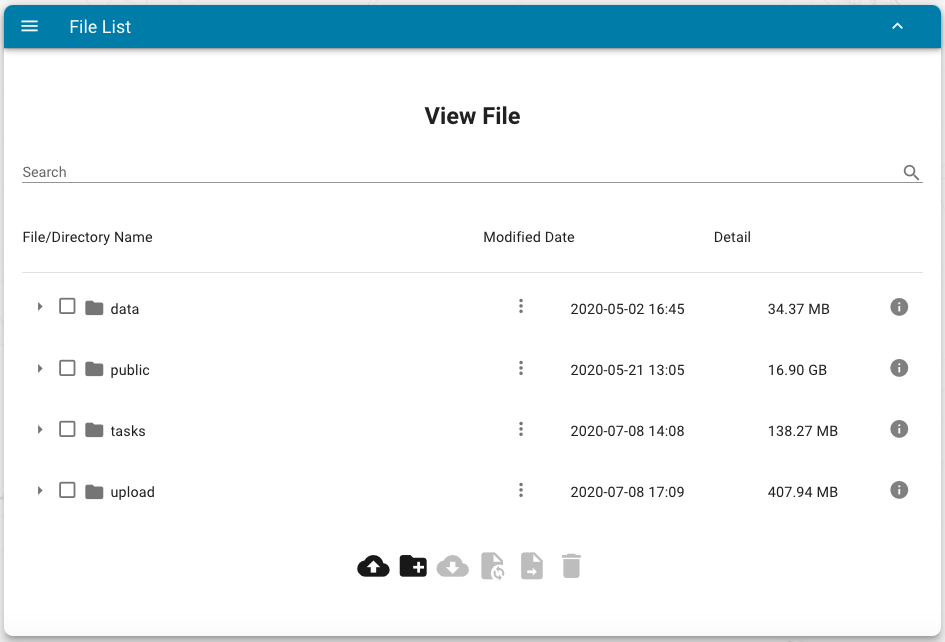
Manager of files
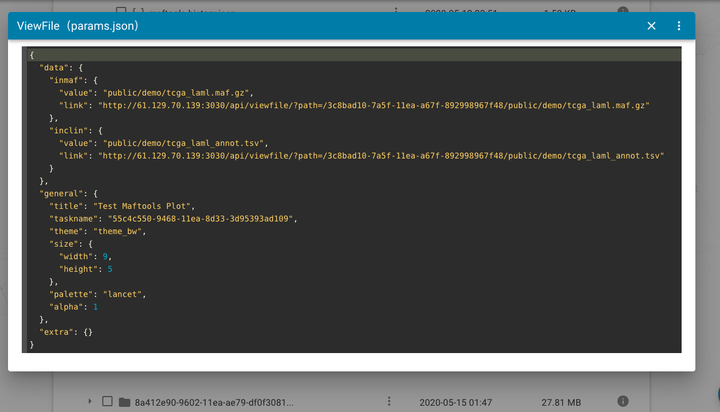
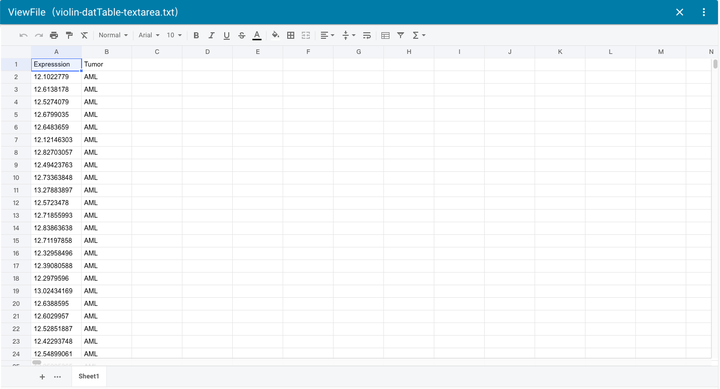
Online view and edit files
Online view and edit files
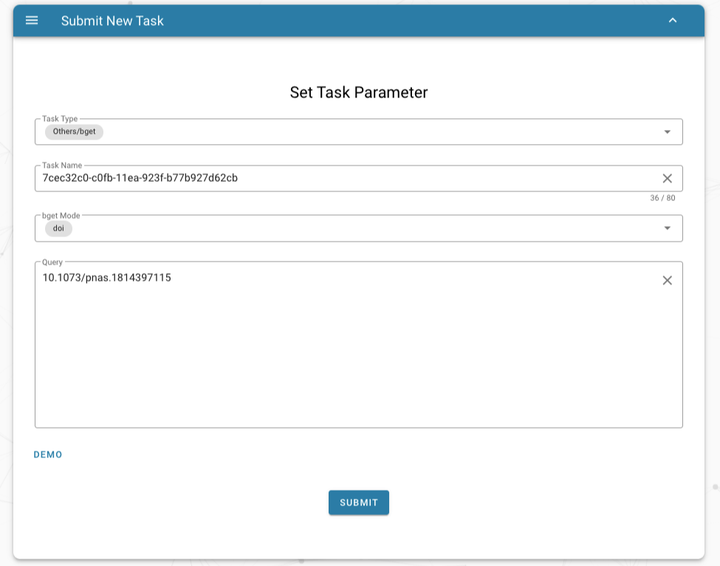
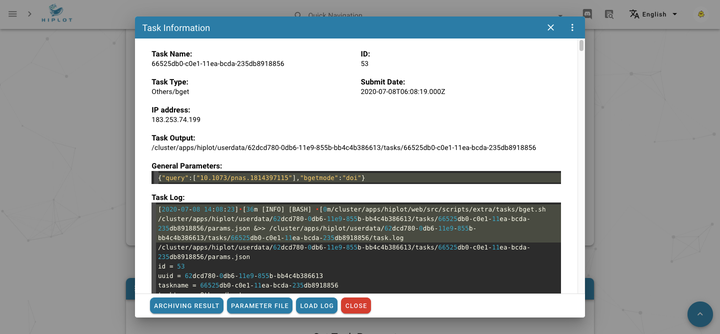
Download publications and supplementary data based on bget
Download publications and supplementary data based on bget
If you want to contribute the Hiplot project, you can fell free to join our community: Discord.
This work is licensed under a Creative Commons Attribution 4.0 International License.