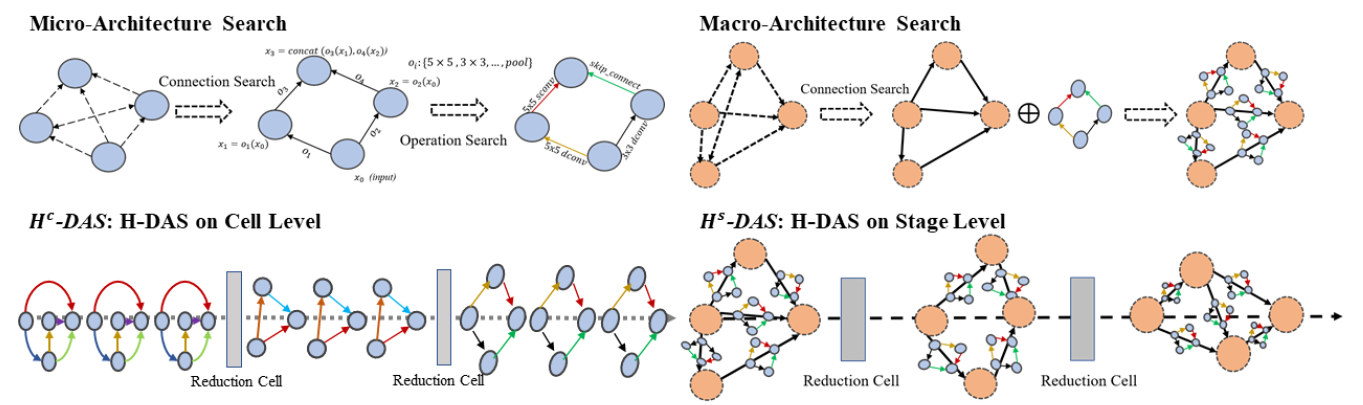
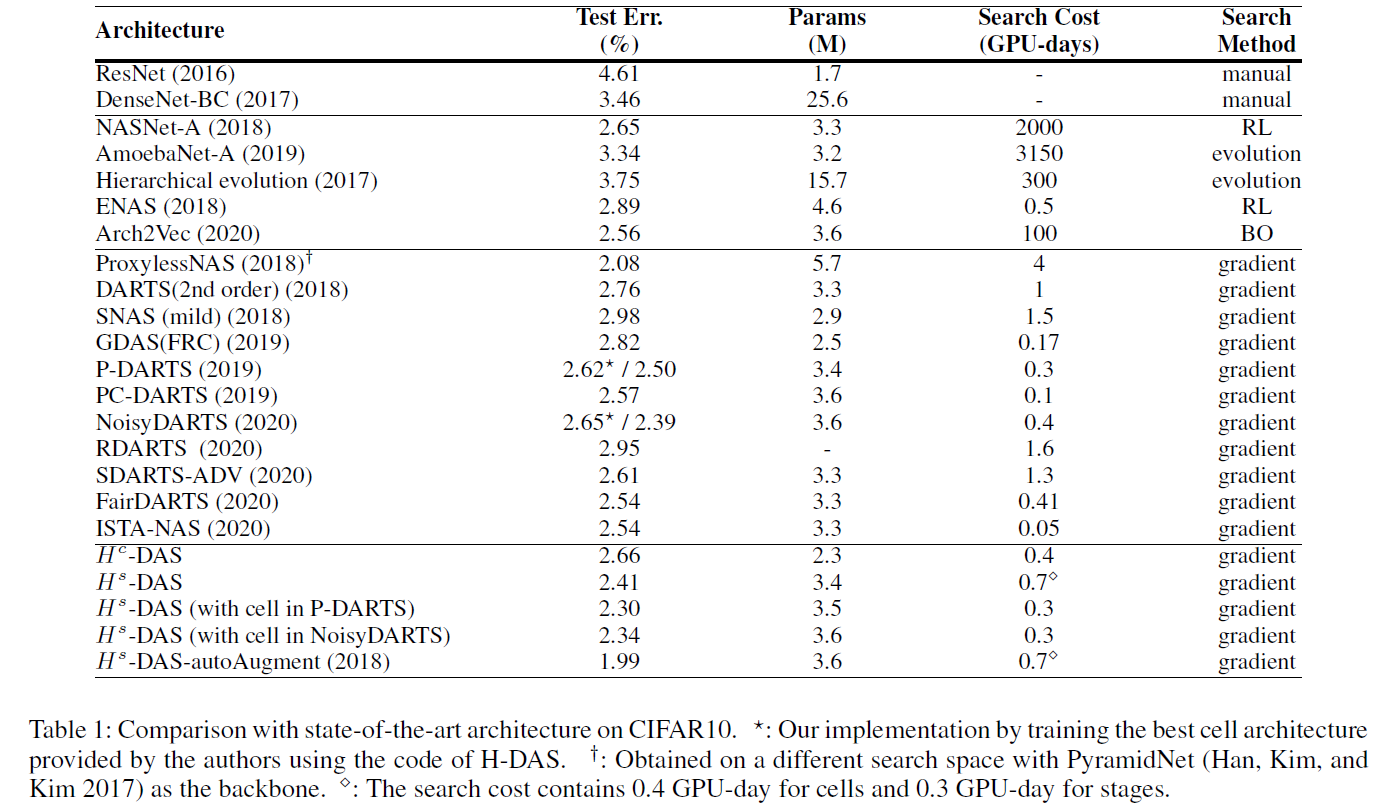
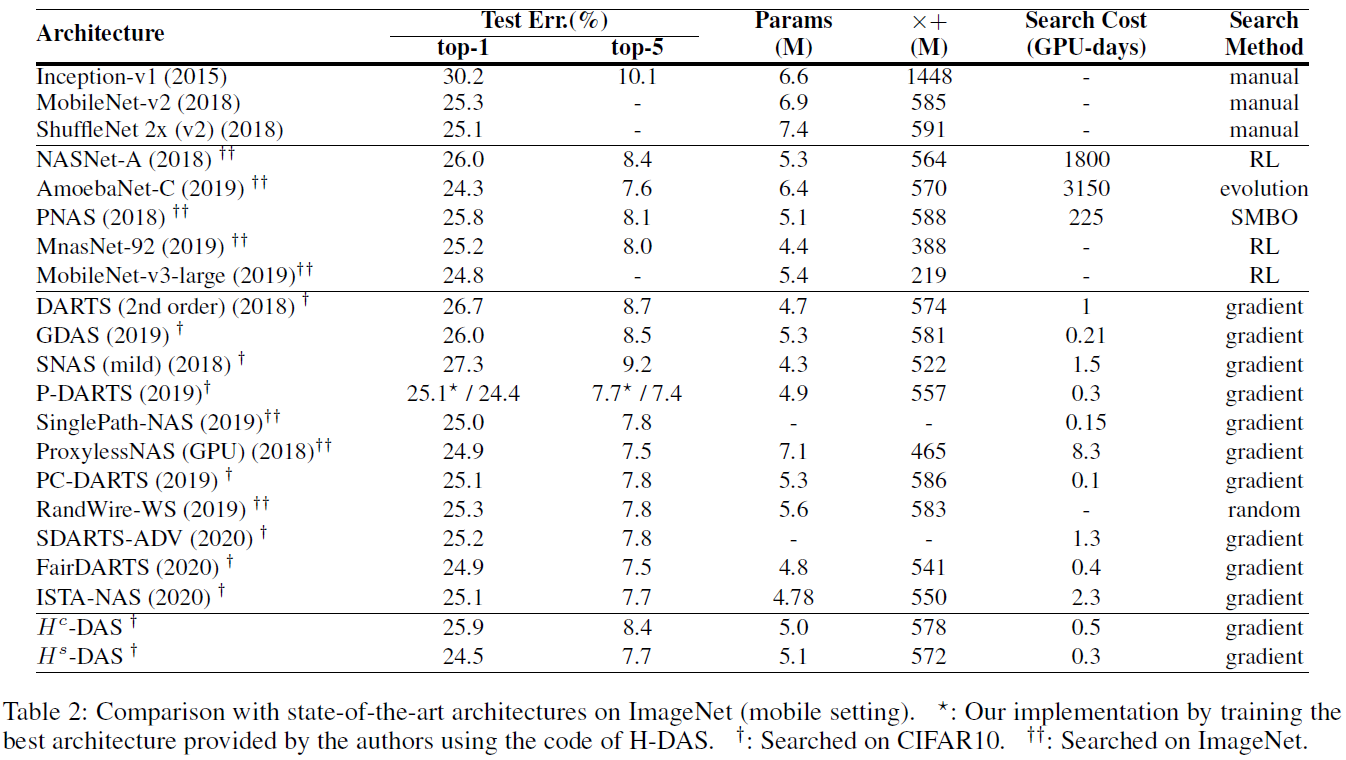
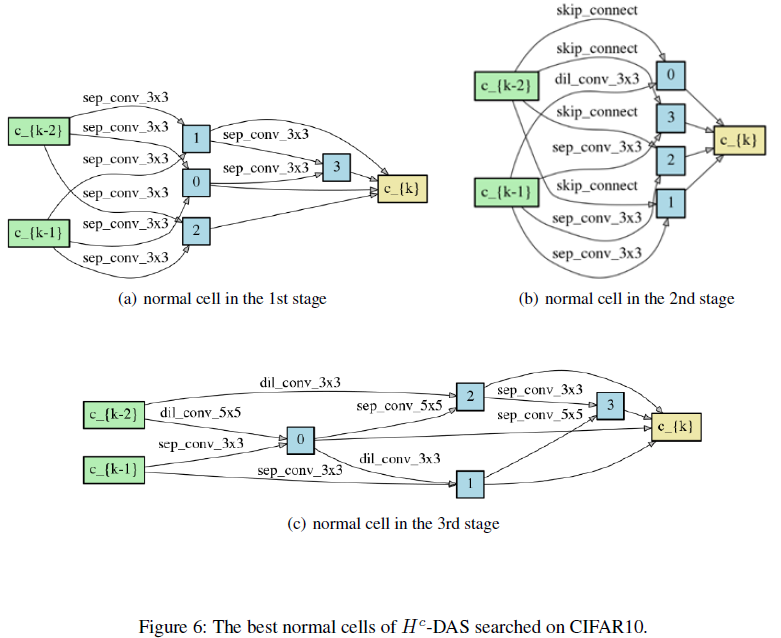
PyTorch implementation of Unchain the Search Space with Hierarchical Differentiable Architecture Search, supporting DDP (DistributedDataParallel) and Apex Amp (Automatic Mixed Precision).
python>=3.6
pytorch>=1.4
tensorboardX
apex
numpy
graphvizWhile CIFAR10 can be automatically downloaded by torchvision, ImageNet needs to be manually downloaded following the instructions here.
Adjust the batch size if out of memory occurs. It depends on your gpu memory size and genotype.
- Search micro-architectures (cell-level structures)
python searchCell_main.py --name cifar10_test --dataset cifar10- Augment micro-architectures (cell-level structures)
python augmentCell_main.py --name Hc-DAS --dataset cifar10 \
--genotype "[Genotype3 searched by 'searchCell_main.py']"- Run the following commands with DistributedDataParallel (ddp_version)
python -m torch.distributed.launch --nproc_per_node=8 \
ddp/augmentCell_main.py --name Hc-DAS --dataset cifar10 \
--genotype "[Genotype3 searched by 'searchCell_main.py']" --dist- Run the following commands to perform a stage-level structure search progress on CIFAR10.
python searchStage_main.py --name macro-cifar10-test \
--w_weight_decay 0.0027 --dataset cifar10 --batch_size 64 \
--workers 0 --genotype "[Genotype searched by DARTS or PDARTS or other Darts-series methods]"- Run the following commands to perform a stage-level structure search progress for ImageNet
python searchStage_ImageNet_main.py --name macro-ImageNet-test \
--dataset cifar10 --batch_size 128 --w_weight_decay 0.0027 \
--workers 16 --genotype "[Genotype searched by DARTS or PDARTS or other Darts-series methods]"- Run the following commands to perform a stage-level structure augment progress on CIFAR10.
python augmentStage_main.py --name Hs-DAS-cifar10-test \
--init_channels 36 --workers 16 --lr 0.025 --batch_size 64 \
--dataset cifar10 --epochs 600 \
--genotype "[Genotype searched by DARTS or PDARTS or other Darts-series methods]" \
--DAG "[Genotype2 searched by 'searchDAG_main.py']"- Run the following commands with DistributedDataParallel (ddp_version)
python -m torch.distributed.launch --nproc_per_node=8 \
ddp/augmentStage_main.py --init_channels 36 --workers 16 \
--name Hs-DAS-cifar10-test --dataset cifar10 \
--genotype "[Genotype searched by DARTS or PDARTS or other Darts-series methods]" \
--DAG "[Genotype2 searched by 'searchDAG_main.py']" --dist- Run the following commands to perform a stage-level structure augment progress on ImageNet.
python augmentStage_ImageNet_main.py --init_channels 39 \
--batch_size 105 --lr 0.1 --workers 16 --dataset imagenet \
--name Hs-DAS_imagenet-test --print_freq 100 --epoch 250 \
--genotype "[Genotype searched by DARTS or PDARTS or other Darts-series methods]" \
--DAG "[Genotype2 searched by 'searchDAG_ImageNet_main.py']"- Run the following commands with DistributedDataParallel (ddp_version)
python -m torch.distributed.launch --nproc_per_node=8 \
ddp/augmentStage_ImageNet_main.py --init_channels 39 \
--workers 16 --name Hs-DAS-imagenet-test --dataset imagenet \
--print_freq 100 --epochs 250 \
--genotype "[Genotype searched by DARTS or PDARTS or other Darts-series methods]" \
--DAG "[Genotype2 searched by 'searchDAG_ImageNet_main.py']" --distWe provide the augmentCell and augmentStage with DistributedDataParallel (ddp_version), but you should change some hyperparameters, such as lr and batch_size.
The search of cell distribution over three stages is performed under a constraint of certain computational complexity.
- Run the following commands to search for the distribution of cells.
python searchDistribution_main.py --name cifar10-changeStage \
--w_weight_decay 0.0027 --dataset cifar10 --batch_size 64 --workers 0 \
--genotype "[Genotype searched by DARTS or PDARTS or other Darts-series methods]"Please cite our paper if this implementation helps your research. BibTex reference is shown in the following.
@inproceedings{liu2021hdas,
title={Unchain the Search Space with Hierarchical Differentiable Architecture Search},
author={Liu, Guanting and Zhong, Yujie and Guo, Sheng and Scott, Matthew R and Huang, Weilin},
booktitle={AAAI},
year={2021}
}For any questions, please fell free to reach:
github@malongtech.comH-DAS is CC-BY-NC 4.0 licensed, as found in the LICENSE file. It is released for academic research / non-commercial use only. If you wish to use for commercial purposes, please contact sales@malongtech.com.