ezancestry
One Thousand Genomes Project -- Population Visualizations using Streamlit
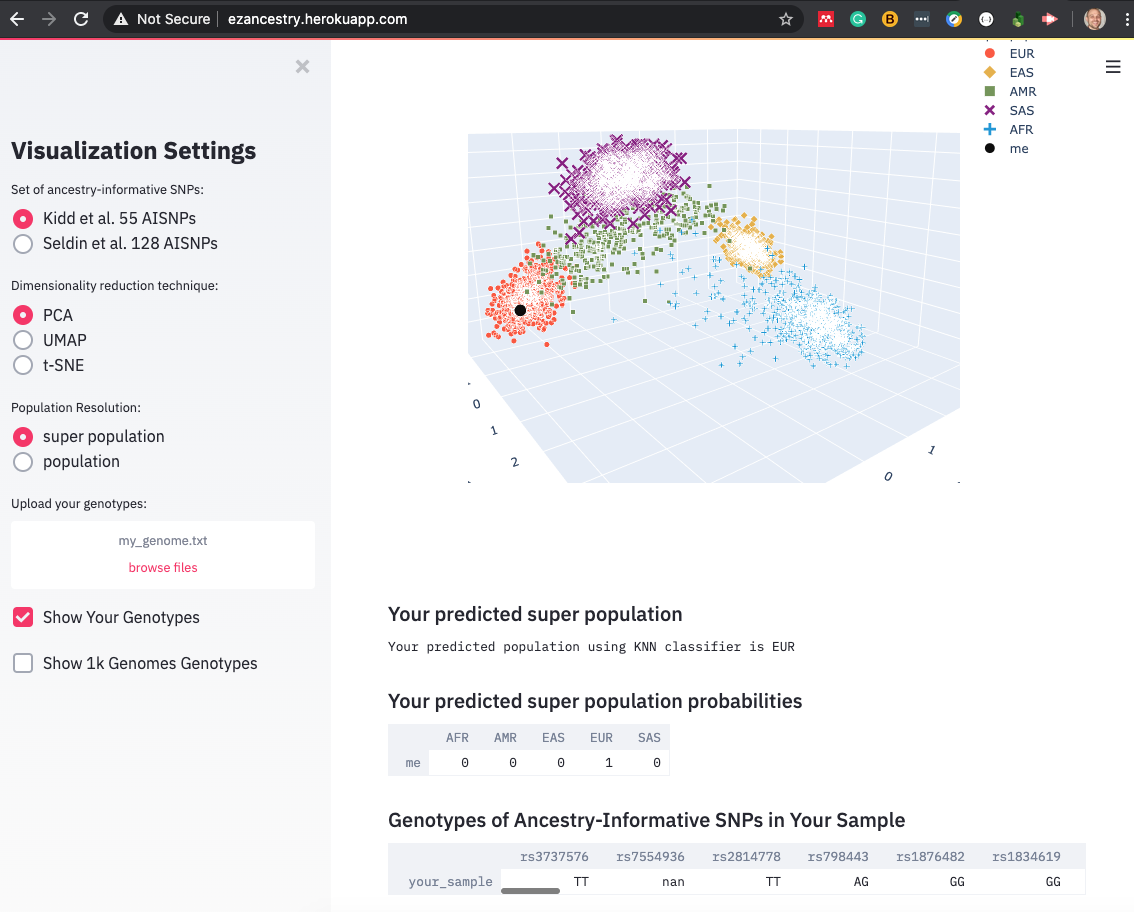
ezancestry is an interactive visualization app built in Python with Streamlit and deployed on Heroku.
Visualize you genotypes at http://ezancestry.herokuapp.com/
Local Installation and Usage
ezancestry can be installed locally:
-
Download
ezancestry -
Change into the
ezancestrydirectory:$ cd ezancestry -
Create a virtual environment:
$ python -m venv .venv -
Activate the virtual environment:
$ source .venv/bin/activate -
Install required packages:
(.venv) $ pip install -r requirements.txtIf issues installing cyvcf2, see here. Setting these environment variables might work (if
opensslinstalled with MacPorts):export LDFLAGS="-L/opt/local/include" export PKG_CONFIG_PATH="/opt/local/include" export CPPFLAGS="-I/opt/local/include" -
Run
ezancestry:(.venv) $ streamlit run app.pyYou will then be presented with a URL to use the app in a browser.
-
Ctrl-C to stop the web server
-
Deactivate the virtual environment:
(.venv) $ deactivate
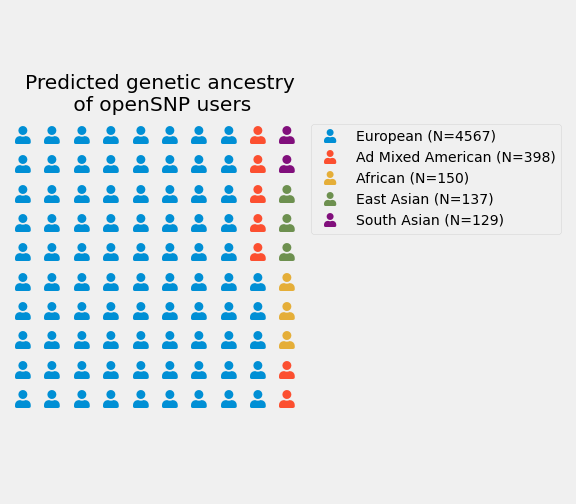
Run ezancestry on openSNP data!
Thanks to Andrew Riha, it's easy to predict the ancestries for all of the openSNP samples:
- Download the data to the
datadirectory. It's a large zipped directory! - Follow steps 1-5 above.
- Run
python opensnp_ancestry.pyand wait for the results.