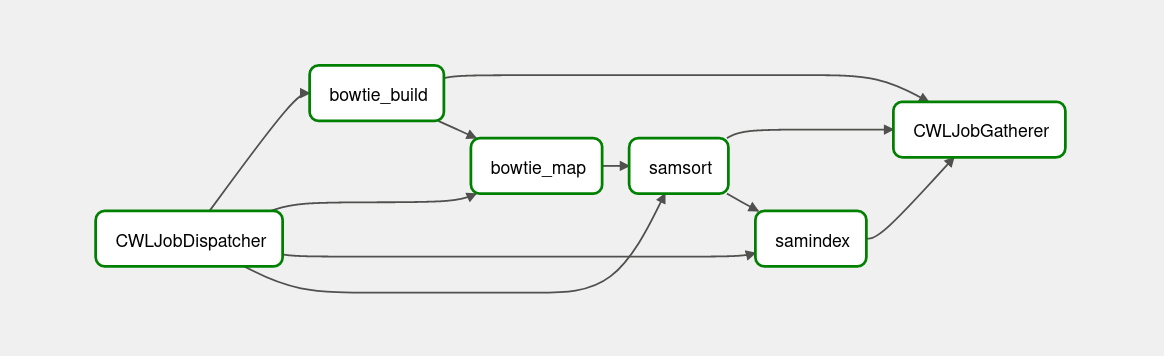
CWL stands for Common Workflow Language, which maybe commonly used in the future because of its strict spec and the ability to cross platform.
CWL should written in Yaml format, due to cross-platofrm compatibility. I think it is not that readable and not easy to write well.
The only flexibilty in CWL is that it allow to write arbitrary expression
e.g. valueFrom: $( inputs.unsort_bam.nameroot + ".bam" ), though it is javascript style.
I generate a fasta file and a pair-end fastq file, then running read-mapping by bowtie2.
In short, we can simlify our pipeline(4 + 1 cwl) into these ipython code (10 lines only).
index = "data/chrx"
fastq = "data/chrx.synthetic"
thread = 30
%alias dk docker run --rm -it -v $PWD:/app -w /app
%dk quay.io/biocontainers/bowtie2:2.4.4--py39hbb4e92a_0 \
bowtie2-build --threads {thread} {index}.fa {index}
%dk quay.io/biocontainers/bowtie2:2.4.4--py39hbb4e92a_0 \
bowtie2 --threads {thread} -x {index} --no-unal -1 {fastq}.read1.fq -2 {fastq}.read2.fq -S {fastq}.bowtie.sam
fastq = fastq + ".bowtie"
%dk quay.io/biocontainers/samtools:1.10--h9402c20_2 \
samtools sort -@{thread} {fastq}.sam -o {fastq}.bam
%dk quay.io/biocontainers/samtools:1.10--h9402c20_2 \
samtools index -@{thread} {fastq}.bam {fastq}.bam.baipip3 install cwltool
rm *.bt2 *.sam *.bam *.bai
# step by step
cwltool --user-space-docker-cmd=podman bowtie2_build.cwl chrx_test.yml
cwltool --user-space-docker-cmd=podman bowtie2_map.cwl chrx_test.yml
cwltool --user-space-docker-cmd=podman samsort.cwl chrx_test.yml
cwltool --user-space-docker-cmd=podman samindex.cwl chrx_test.yml
# Run all steps in once
cwltool --user-space-docker-cmd=podman pipeline.cwl chrx.ymlAirflow is a visualization & DAG manage tool
# install (cwl-airflow will install airflow too)
pip3 install cwl-airflow
# init airflow
export AIRFLOW_HOME=$PWD/airflow/
airflow db init
airflow users create -e linnil1 -f linnil1 -l linnil1 -r Admin -u linnil1 -p password
# add my dags and copy input files
mkdir airflow/dags/
sed -e "s|\$PWD|$PWD|g" airflow/bowtie2_cwl.py > airflow/dags/bowtie2_cwl.py
cp -r data/ airflow/cwl_inputs_folder
# run
airflow webserver
# Run this in another window
export AIRFLOW_HOME=$PWD/airflow/
airflow scheduler
# Run this in another window
export AIRFLOW_HOME=$PWD/airflow/
cwl-airflow init
cwl-airflow api
# Open browser localhost:8080
# uername: linnil1
# password: password
# Find bowtie2_cwl
# **Unpause DAG**
# **Trigger with config**
# Paste the output of this
python3 -c "import json; import yaml; print(json.dumps({'job': yaml.load(open('chrx.yml'))}))"
# {
# "job": {
# "reference_fasta": {"class": "File", "path": "data/chrx.fa"},
# "fastq_1": {"class": "File", "path": "data/chrx.synthetic.read1.fq"},
# "fastq_2": {"class": "File", "path": "data/chrx.synthetic.read2.fq"},
# "threads": 10}
# } - Suffix name
- airflow
- Log
- Set fastq paired-end reads as secondaryFiles
- CWL scatter
- put file into correct folder, not flatten structure (Partially solved)
- Skip if output exist (No you cannot)
What is suffix name
Put every steps' short_description into suffix
.
├── chrx.fa
├── chrx.1.bt2
├── chrx.2.bt2
├── chrx.3.bt2
├── chrx.4.bt2
├── chrx.rev.1.bt2
├── chrx.rev.2.bt2
├── chrx.synthetic.read1.fq
├── chrx.synthetic.read2.fq
├── chrx.synthetic.bowtie.bam
└── chrx.synthetic.bowtie.bam.bai
Why don't write the default arguments with expression like below?
Because cwl doesn't support this syntax
inputs:
output_bam_file:
type: string
inputBinding:
prefix: -o
default: $( inputs.unsort_bam.nameroot + ".bam" )Thus, arguments part is used instead.
arguments:
- valueFrom: $( inputs.unsort_bam.nameroot + ".bam" )
prefix: -ologging
I know there is a way to save the log in each cwl, but Addin stdout and stderr for each steps is tidious ().
bowtie2_build.cwl
outputs:
stdout:
type: stdout
stderr:
type: stderr
stdout: $( inputs.reference_fasta.nameroot + ".bt2.log" )
stderr: $( inputs.reference_fasta.nameroot + ".bt2.err" )pipeline.cwl
steps:
bowtie_build:
...
out: [bowtie2_index, stdout, stderr]
outputs:
# log (really stupid)
bowtie_build_log:
type: File
outputSource: bowtie_build/stdout
bowtie_build_err:
type: File
outputSource: bowtie_build/stderrbut I think it's tidious to write log, so I patch the code
diff --git a/cwl_airflow/utilities/cwl.py b/cwl_airflow/utilities/cwl.py
index 7b1ea18..ea06aa5 100644
@@ -652,12 +653,15 @@ def execute_workflow_step(
if need_to_run(workflow_data, job_data, task_id):
skipped = False
_stderr = sys.stderr # to trick the logger
- sys.stderr = sys.__stderr__
+ sys.stderr = open("/tmp/tmp_file_to_save_stderr", "w")
step_outputs, step_status = executor(
workflow_data,
job_data,
RuntimeContext(default_cwl_args)
)
+ sys.stderr.close()
+ logging.info(open("/tmp/tmp_file_to_save_stderr").read())
sys.stderr = _stderrThe dependency graph of cwl
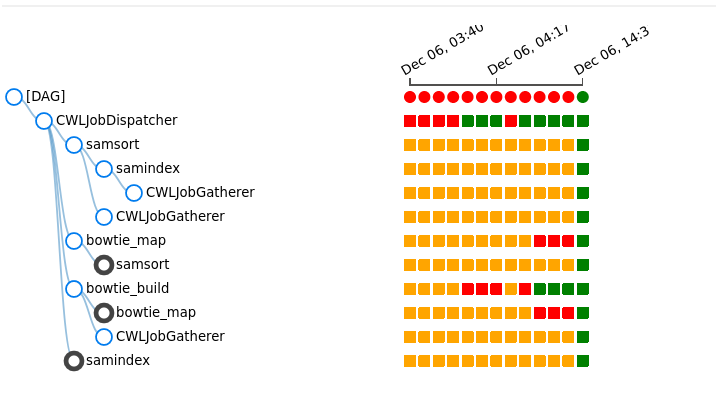
The status of cwl of each triggers
Output files
[linnil1@linnil1 cwl]$ ls airflow/cwl_outputs_folder/bowtie2_cwl/manual__2021-12-07T06_56_33.308666_00_00/
chrx.1.bt2 chrx.3.bt2 chrx.rev.1.bt2 chrx.synthetic.bowtie.bam workflow_report.json
chrx.2.bt2 chrx.4.bt2 chrx.rev.2.bt2 chrx.synthetic.bowtie.bam.bai
Logs
airflow/logs/bowtie2_cwl/bowtie_map/2021-12-09T08\:21\:30.630135+00\:00/1.log
[2021-12-09 14:03:31,423] {workflow_job.py:787} INFO - [workflow ] start
[2021-12-09 14:03:31,426] {workflow_job.py:626} INFO - [workflow ] starting step bowtie_map
[2021-12-09 14:03:31,427] {workflow_job.py:74} INFO - [step bowtie_map] start
[2021-12-09 14:03:31,697] {job.py:752} INFO - ['podman', 'pull', 'quay.io/biocontainers/bowtie2:2.4.4--py39hbb4e92a_0']
[2021-12-09 14:03:35,983] {job.py:259} INFO - [job bowtie_map] /home/linnil1/airflow/cwl_tmp_folder/bowtie2_cwl_manual__2021-12-09T06_02_56.509681_00_00_nh4jkmbh/bowtie_map/bowtie_map_step_cache/lfec60jp$ podman \
run \
--volume=/home/linnil1/airflow/cwl_tmp_folder/bowtie2_cwl_manual__2021-12-09T06_02_56.509681_00_00_nh4jkmbh/bowtie_map/bowtie_map_step_cache/lfec60jp:/EevVJN \
--volume=/home/linnil1/airflow/cwl_tmp_folder/bowtie2_cwl_manual__2021-12-09T06_02_56.509681_00_00_nh4jkmbh/bowtie_map/bowtie_map_step_cache/7nvwhs2j:/tmp \
--volume=/home/linnil1/airflow/cwl_inputs_folder/data/chrx.synthetic.read1.fq:/var/lib/stg84c69277-4631-4adf-a0d6-dfbc86b4219a/chrx.synthetic.read1.fq \
--volume=/home/linnil1/airflow/cwl_inputs_folder/data/chrx.synthetic.read2.fq:/var/lib/stg6072d412-5471-48ca-b12d-d799f6d010ab/chrx.synthetic.read2.fq \
--volume=/home/linnil1/airflow/cwl_tmp_folder/bowtie2_cwl_manual__2021-12-09T06_02_56.509681_00_00_nh4jkmbh/bowtie_build/bowtie_build_step_outputs/chrx.1.bt2:/var/lib/stg62f381c5-9c43-4baa-a91e-3f39c250f8dd/chrx.1.bt2 \
--volume=/home/linnil1/airflow/cwl_tmp_folder/bowtie2_cwl_manual__2021-12-09T06_02_56.509681_00_00_nh4jkmbh/bowtie_build/bowtie_build_step_outputs/chrx.2.bt2:/var/lib/stg62f381c5-9c43-4baa-a91e-3f39c250f8dd/chrx.2.bt2 \
--volume=/home/linnil1/airflow/cwl_tmp_folder/bowtie2_cwl_manual__2021-12-09T06_02_56.509681_00_00_nh4jkmbh/bowtie_build/bowtie_build_step_outputs/chrx.3.bt2:/var/lib/stg62f381c5-9c43-4baa-a91e-3f39c250f8dd/chrx.3.bt2 \
--volume=/home/linnil1/airflow/cwl_tmp_folder/bowtie2_cwl_manual__2021-12-09T06_02_56.509681_00_00_nh4jkmbh/bowtie_build/bowtie_build_step_outputs/chrx.4.bt2:/var/lib/stg62f381c5-9c43-4baa-a91e-3f39c250f8dd/chrx.4.bt2 \
--volume=/home/linnil1/airflow/cwl_tmp_folder/bowtie2_cwl_manual__2021-12-09T06_02_56.509681_00_00_nh4jkmbh/bowtie_build/bowtie_build_step_outputs/chrx.rev.1.bt2:/var/lib/stg62f381c5-9c43-4baa-a91e-3f39c250f8dd/chrx.rev.1.bt2 \
--volume=/home/linnil1/airflow/cwl_tmp_folder/bowtie2_cwl_manual__2021-12-09T06_02_56.509681_00_00_nh4jkmbh/bowtie_build/bowtie_build_step_outputs/chrx.rev.2.bt2:/var/lib/stg62f381c5-9c43-4baa-a91e-3f39c250f8dd/chrx.rev.2.bt2 \
--workdir=/EevVJN \
--rm \
--env=TMPDIR=/tmp \
--env=HOME=/EevVJN \
quay.io/biocontainers/bowtie2:2.4.4--py39hbb4e92a_0 \
bowtie2 \
--no-unal \
-S \
chrx.synthetic.bowtie.sam \
-1 \
/var/lib/stg84c69277-4631-4adf-a0d6-dfbc86b4219a/chrx.synthetic.read1.fq \
-2 \
/var/lib/stg6072d412-5471-48ca-b12d-d799f6d010ab/chrx.synthetic.read2.fq \
-x \
/var/lib/stg62f381c5-9c43-4baa-a91e-3f39c250f8dd/chrx \
--threads \
8
[2021-12-09 14:03:37,550] {job.py:524} INFO - [job bowtie_map] Max memory used: 138MiB
[2021-12-09 14:03:37,562] {job.py:403} INFO - [job bowtie_map] completed success
[2021-12-09 14:03:37,562] {workflow_job.py:587} INFO - [step bowtie_map] completed success
[2021-12-09 14:03:37,563] {workflow_job.py:549} INFO - [workflow ] completed success
[2021-12-09 14:03:37,567] {cwl.py:666} INFO - 2bedd08e8057a32b6411dabae5828af2459529a4394bc89a9c70e33a753e7e5e
1650 reads; of these:
1650 (100.00%) were paired; of these:
591 (35.82%) aligned concordantly 0 times
682 (41.33%) aligned concordantly exactly 1 time
377 (22.85%) aligned concordantly >1 times
----
591 pairs aligned concordantly 0 times; of these:
543 (91.88%) aligned discordantly 1 time
----
48 pairs aligned 0 times concordantly or discordantly; of these:
96 mates make up the pairs; of these:
0 (0.00%) aligned 0 times
48 (50.00%) aligned exactly 1 time
48 (50.00%) aligned >1 times
100.00% overall alignment rate
[2021-12-09 14:03:37,585] {taskinstance.py:1212} INFO - Marking task as SUCCESS. dag_id=bowtie2_cwl, task_id=bowtie_map, execution_date=20211209T060256, start_date=20211209T060325, end_date=20211209T060337
[2021-12-09 14:03:37,640] {local_task_job.py:151} INFO - Task exited with return code 0
[2021-12-09 14:03:37,682] {local_task_job.py:261} INFO - 1 downstream tasks scheduled from follow-on schedule check
Folder
It's not that easy than I though, I need to add folder manually.
hints:
InitialWorkDirRequirement:
listing:
- entry: $( inputs.output_folder )
arguments:
- valueFrom: $( inputs.output_folder.path + "/" + inputs.fastq_1.basename.slice(0, -9) + ".bowtie.sam" )
prefix: -S
inputs:
output_folder:
type: Directory
outputs:
bowtie_output_sam_tmp:
type: File
outputBinding:
glob: $( inputs.output_folder.path + "/" + inputs.fastq_1.basename.slice(0, -9) + ".bowtie.sam" )- CWL spec
- cwltool
- cwl-airflow
- bowtie2
- samtools
- podman
MIT