The goal of single cell Genome and Epigenome by Transposases sequencing (scGET-seq) is to discriminate between accessible and compacted chromatin regions within each single cell. The discrimination of chromatin accessibility relies on two different transposases: transposase-5 binds to the accessible chromatin (tn5) and transposase-H, a chimeric form of tn5 (tnH), which recognizes the compacted chromatin.
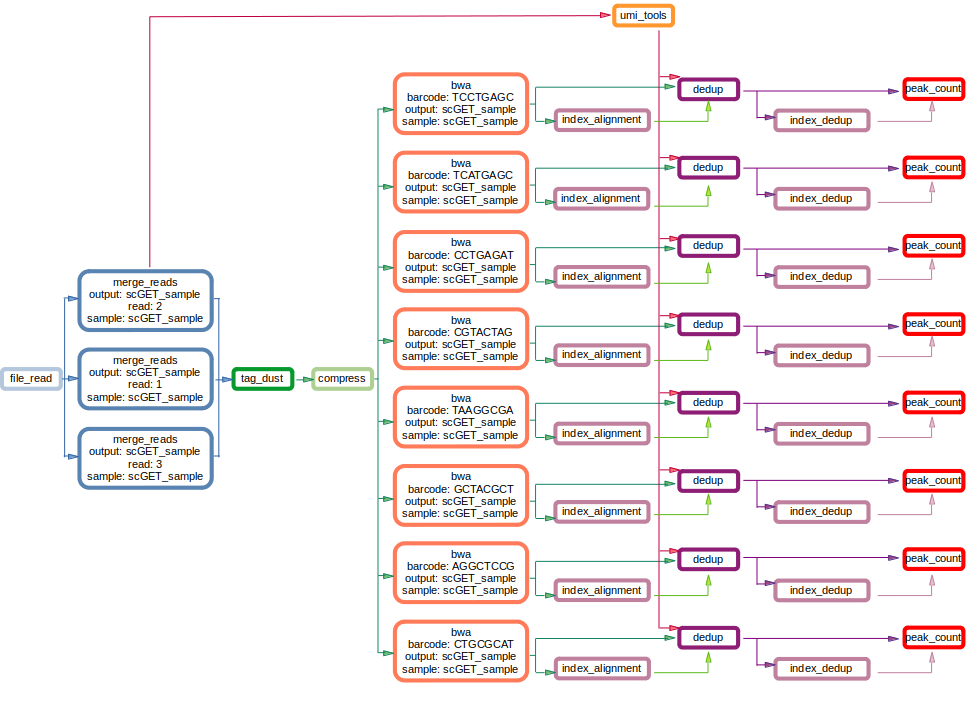
scGET architecture is built using Snakemake: a workflow management system, which guarantees the possibility to parallelize independent jobs. scGET workflow is described by the image below: starting from sequenced FASTQ files, scGET will generate an AnnData object where tn5 matrix and tnh matrix are present as two different layers.
First, scGET repository must be cloned:
git clone --recursive https://github.com/leomorelli/scGET.git
Before getting your hands dirty with scGET analyses, it is necessary to create a suitable conda environment. However, some packages cannot be installed, using conda. Therefore, we have designed a 4-step process, allowing an easy and quick generation of the scget environment.
- The conda environment can be automatically generated, thanks to the scget.yaml file:
conda env create -f scget.yaml
conda activate scget
TagDustpackage must be installed, after the activation of the scget environment. First, the package must be downloaded and compiled; second, from the tagdust directory,the binary tagdust file can be copied in the scget environment:
wget https://sourceforge.net/projects/tagdust/files/tagdust-2.33.tar.gz
tar -zxvf tagdust-2.33.tar.gz
cd tagdust-2.33
./configure
make
make check
cp ./src/tagdust $CONDA_PREFIX/bin
- Similarly, also
samtoolsmust be installed:- git repositories of
samtoolsandhtslibmust be cloned htslibmust be compiled and installedsamtoolsmust be compiled and installed
- git repositories of
git clone https://github.com/samtools/samtools.git
git clone https://github.com/samtools/htslib.git
cd htslib
autoreconf -i
git submodule update --init --recursive
./configure --prefix=$CONDA_PREFIX
make
make install
cd samtools
autoheader
autoconf -Wno-syntax
./configure --prefix=$CONDA_PREFIX --without-curses
make
make install
scatACCrepository should be automatically retrieved within the current repository, otherwise it must be cloned from github:
git clone https://github.com/dawe/scatACC.git
In order to perform the analysis through slurm, it may be useful to check if screen package has already been installed:
screen --version
Output (example):
Screen version 4.08.00 (GNU) 05-Feb-20
If screen has not been installed yet, it could be easily installed via sudo:
sudo apt update
sudo apt install screen
Although scGET can be used locally, it is optimized to work on a cluster, managed by Slurm workload manager.
- Inside
${HOME}/.config, a series of nested directories should be created, such that you obtain the following path${HOME}/.config/snakemake/slurm. Inside the slurm folder, a config.yaml file can be generated:
mkdir -p ${HOME}/.config/snakemake/slurm
cd ${HOME}/.config/snakemake/slurm
vi config.yaml
- After that, the config.yaml file must be compiled as explained below (remember to update the queue name specified by the
-poption and yourmail-user):
jobs: 38
cluster: "sbatch --mem={resources.mem_mb} -c {resources.cpus} --job-name {rule}.smk -o {OUTPUT_PATH}/logs_slurm/{rule}_%j.o -e {OUTPUT_PATH}/logs_slurm/{rule}_%j.e --mail-type=FAIL --mail-user=user@mail.com"
default-resources: [cpus=1, mem_mb=5000]
resources: [cpus=40, mem_mb=60000]
restart-times: 3
use-conda: true
The path for scatACC directory (should be within the current directory), together with the path for the genome and the bed_file must be clarified in the config.yaml present in the scGET folder.
EXAMPLE:
Let's assume that the scGET directory is located in our home directory (${HOME}/scGET); scatACC directory is then in a directory ${HOME}/scGET/scatACC); on the other hand, the genome file (hg38.fa), lays in the "references" directory (${HOME}/references/hg.38), together with the bed_file (${HOME}/references/hg385kbin.bed):
- First, you should open the
config.yamlfile, in thescGETdirectory:
cd ${HOME}/scGET
vi config.yaml
Output:
sample: ''
reads: [1,2,3]
barcodes: {'tn5':['CGTACTAG','TCCTGAGC','TCATGAGC','CCTGAGAT'],'tnh':['TAAGGCGA','GCTACGCT','AGGCTCCG','CTGCGCAT']}
genome: ${HOME}/genome.fa
bed_file: ${HOME}/genome.bed
threads: 8
cell_number: 5000
scatacc_path: '${HOME}/scGET/scatACC'
input_path: ''
input_list: ''
output_path: ''
- After that, we must modify the field
scatacc_path, specifying our actual scatACC path, the fieldgenome, clarifying the genome path with the genome file name and the fieldbed_filewith the path for the bed file:
Output:
sample: ''
reads: [1,2,3]
barcodes: {'tn5':['CGTACTAG','TCCTGAGC','TCATGAGC','CCTGAGAT'],'tnh':['TAAGGCGA','GCTACGCT','AGGCTCCG','CTGCGCAT']}
genome: ${HOME}/references/hg38.fa
bed_file: ${HOME}/references/hg385kbin.bed
threads: 8
cell_number: 5000
scatacc_path: '${HOME}/scGET/scatACC'
input_path: ''
input_list: ''
output_path: ''
N.B. the REFERENCE GENOME must be INDEXED before the analysis
If the genome has not been indexed yet, you can make up for this in three steps:
- Activate the scget conda environment
- Open the directory where the reference genome is stored
- Index the genome, using samtools library
conda activate scget
cd ${HOME}/references
bwa index hg38.fa
Two inputs are mandatory to start the scGET analisys:
- The path for fastq input files
- A .txt file, listing names of the files ready to be analyzed
EXAMPLE:
Let's assume that fastq files are stored in ${HOME}/files/samples directory: ${HOME}/files/samples represents the input path; while names of files inside ${HOME}/files/samples directory represent the content of the .txt file, we must create.
ls ${HOME}/files/samples
Output:
sample_S1_L001_R1_001.fastq.gz
sample_S1_L001_R2_001.fastq.gz
sample_S1_L001_R3_001.fastq.gz
sample_S1_L002_R1_001.fastq.gz
sample_S1_L002_R2_001.fastq.gz
sample_S1_L002_R3_001.fastq.gz
From the output above, it easy to understand which read number corresponds to each file (R1, R2 and R3). The .txt file, must be built as follow:
- Each line corresponding to a file name
- Next to the file name, the read number should be clarified
- Finally, the sample name must be indicated next to the read number. This step allows the simultanous analysis of different samples. -> file.fq.gz | read_n° | sample_name
EXAMPLE:
vi input_info.txt
After that, it must be modified as explained below:
sample_S1_L001_R1_001.fastq.gz 1 S1
sample_S1_L001_R2_001.fastq.gz 2 S1
sample_S1_L001_R3_001.fastq.gz 3 S1
sample_S1_L002_R1_001.fastq.gz 1 S1
sample_S1_L002_R2_001.fastq.gz 2 S1
sample_S1_L002_R3_001.fastq.gz 3 S1
sample_S2_L001_R1_001.fastq.gz 1 S2
sample_S2_L001_R2_001.fastq.gz 2 S2
sample_S2_L001_R3_001.fastq.gz 3 S2
sample_S2_L002_R1_001.fastq.gz 1 S2
sample_S2_L002_R2_001.fastq.gz 2 S2
sample_S2_L002_R3_001.fastq.gz 3 S2
Now it's time to start the analysis! It is important to remember that the scGET analysis must be performed from the scGET directory or from a directory in which the Snakefile, the config.yaml and the scripts files are copied. Therefore, before starting the workflow, you should reach the scGET directory and activate the scGET environment.
cd ${HOME}/scGET
conda activate scget
In order to start with scGET analysis, you must run the following command, specifying the input_path, the output_path and the input_list generated above:
snakemake --cores 8 --config input_path=/home/files/experiment_test output_path=/home/results input_list=input_file.txt --profile slurm
Once scGET analysis is finished results files as well as log files are generated and stored in the output directory:
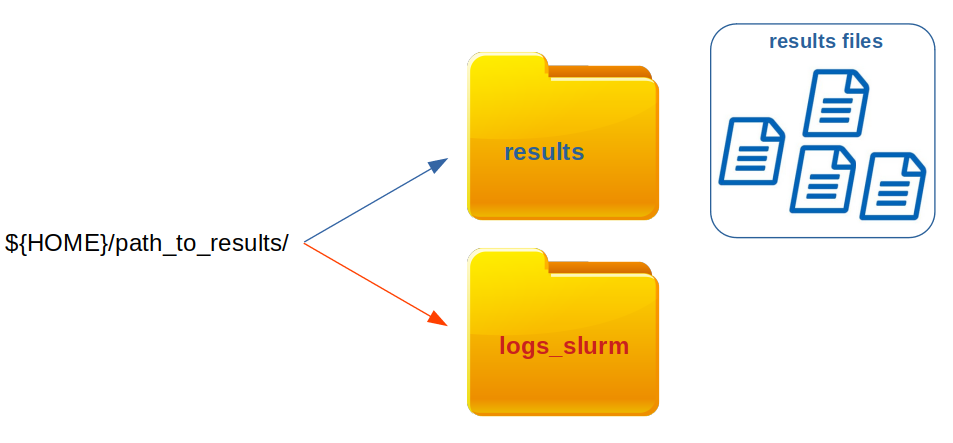
- Results files are stored in a directory named after the sample name
- Log files are stored in the
logs_slurmdirectory, located in the directory, indicated by theoutput_path
The location of results directory is indicated by the parameter output_path.
N.B.
If you need to dig more into scGET settings, you can find more info about scGET usage in the advanced.md file.