🌿 Cassvana Leaf Disease Classification
Kaggle Competition: https://www.kaggle.com/c/cassava-leaf-disease-classification
Dataset
- Train set: ~26,000 images (21367 images of the 2020 contest was merged with 500 images from the 2019 contest).
- Test set: ~15,000 images.
- Public test: 31% of the test set.
- Private test: 69% of the test set.
The dataset is imbalanced with 5 labels:
- Cassava Bacterial Blight (CBB)
- Cassava Brown Streak Disease (CBSD)
- Cassava Green Mottle (CGM)
- Cassava Mosaic Disease (CMD)
- Healthy
| Original data images |
| After albumentations |
Requirements
Python >= 3.8. Run this command to install all the dependencies:
pip install -r requirements.txt
Directories Structures
this repo
└─── train_images
│ └─── ***.png # Dataset folder
└─── test_images
│ └─── ***.png
|
└─── configs # Config folder
│ └─── train.yaml
│ └─── test.yaml
│ └─── config.py
|
└─── csv # labels folder
│ └─── folds
│ └─── fold_train.csv
│ └─── fold_val.csv
│
└─── loggers # experiments folder
│ └─── runs
│ └─── loss_fold
| └─── acc_fold
└─── weights # experiments folder
│ └─── model_name1.pth
| └─── ...
|
|
train.py
test.py
Edit YAML
Full explanation on each YAML file
Training Steps
- Download and unzip dataset from https://www.kaggle.com/c/cassava-leaf-disease-classification/data
- Run this command to split train.csv using KFold. A folder name 'csv' will be created with different k-folds
python utils/tools/split_kfold.py --csv=train.csv --seed=42 --out=csv --n_splits=5 --shuffle=True
- Run this command on terminal or colaboratory (change if needed)
tensorboard --logdir='./loggers/runs'
- Run this command and fine-tune on parameters for fully train observation (Require change)
python train.py --config=config_name --resume=weight_path --print_per_iters=100 --gradcam_visualization
- The model weights will be saved automatically in the 'weights' folder
Inference
Run this command to generate predictions and submission file (Require fine-tune inside)
python test.py --config=test
Result
I have trained on Efficientnet-b6, EfficientNet-b1 and ViT. Here are the results:
- The result from Efficientnet-b6 is not quite good, accuracy just between 0.7-0.8 before Early Stopping.
- The result from Efficientnet-b1 and ViT are good enough: about 0.87x each.
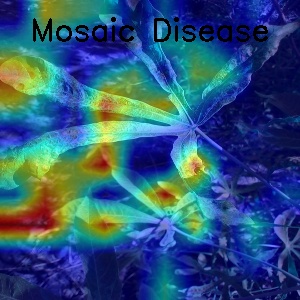
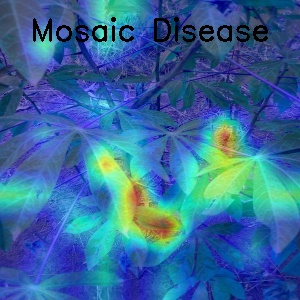
- Some visualization for Mosaic Disease with GradCam
To-do list:
- Multi-GPU support (nn.DataParallel)
- GradCAM vizualization
- Gradient Accumulation
- Mixed precision
- Stratified KFold splitting
- Inference with Ensemble Model technique and TTA
- Metrics: Accuracy, Balanced Accuracy, F1-Score
- Losses: Focal Loss, SmoothCrossEntropy Loss
- Optimizer: AdamW, SGD. Adas, SAM (not debug yet)
- Scheduler: ReduceLROnPlateau, CosineAnnealingWarmRestarts
- Usable Models: Vit, EfficientNet, Resnext, Densenet
- Early Stopping on training
Reference:
Make sure to give them a star
- Template from: https://github.com/kaylode/custom-template
- timm models from https://github.com/rwightman/pytorch-image-models