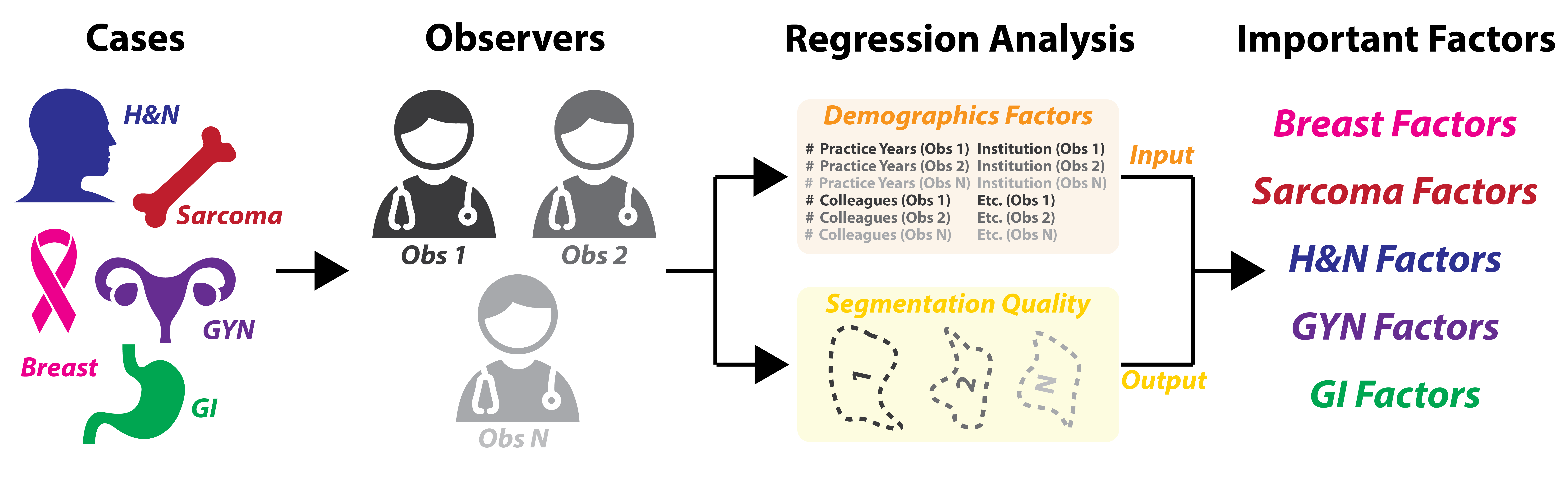
Determining The Role Of Radiation Oncologist Demographic Factors On Segmentation Quality: Insights From A Crowd-Sourced Challenge Using Bayesian Estimation
Pre-print available at: https://www.medrxiv.org/content/10.1101/2023.08.30.23294786v2. Corresponding image sets used for this project are avaliable on Figshare (data doi: 10.6084/m9.figshare.21074182). Data descriptor located at: https://www.medrxiv.org/content/10.1101/2022.10.05.22280672v1. All data has been anonymized to remove patient PHI. The CSV files generated as data for this specific project can also be found on Figshare (doi:10.6084/m9.figshare.24021591).
Jupyter notebook of scripts to generate CSV files for regression analysis (Base_file_generation.ipynb).
Jupyter notebook of scripts to perform statistical analysis in paper (Statistical_analysis_code.ipynb).
Folders for various output files (bambi_binary_bayesian_regression_outputs, csv_files, descriptive_stats, plot_outputs).
For ease of code reproduction, we have generated a YAML file with the conda virtual environment parameters that were used to run the most recent version of the code (C3RO_regression.yml). We used conda version 4.12.0. Unfortunately, some of the conda packages we used implemented Windows-specific build hash strings, so exact decimal-level reproducibility precision cannot be ensured if you try to install the environment on a different OS. Also please note, one of the required libraries for metric calculations (surface_distance) will require the user to pull a GitHub repo onto their local folders. See instructions here:https://github.com/google-deepmind/surface-distance.
To install a new conda environment on your system and ensure you can access the kernel in Jupyter, use the following commands:
- conda env create -f C3RO_regression.yml -n C3RO_regression
- python -m ipython install --user --name C3RO_regression --display-name "C3RO_regression"
arviz version 0.15.1.
bambi version 0.12.0.
pymc version 5.6.1.
SimpleITK version 2.3.0.
Numpy version 1.24.4.
Pandas version 2.0.3.
Surface-Distance-Based-Measures version 0.1.
Matplotlib version 3.7.2.
Seaborn version 0.12.2.