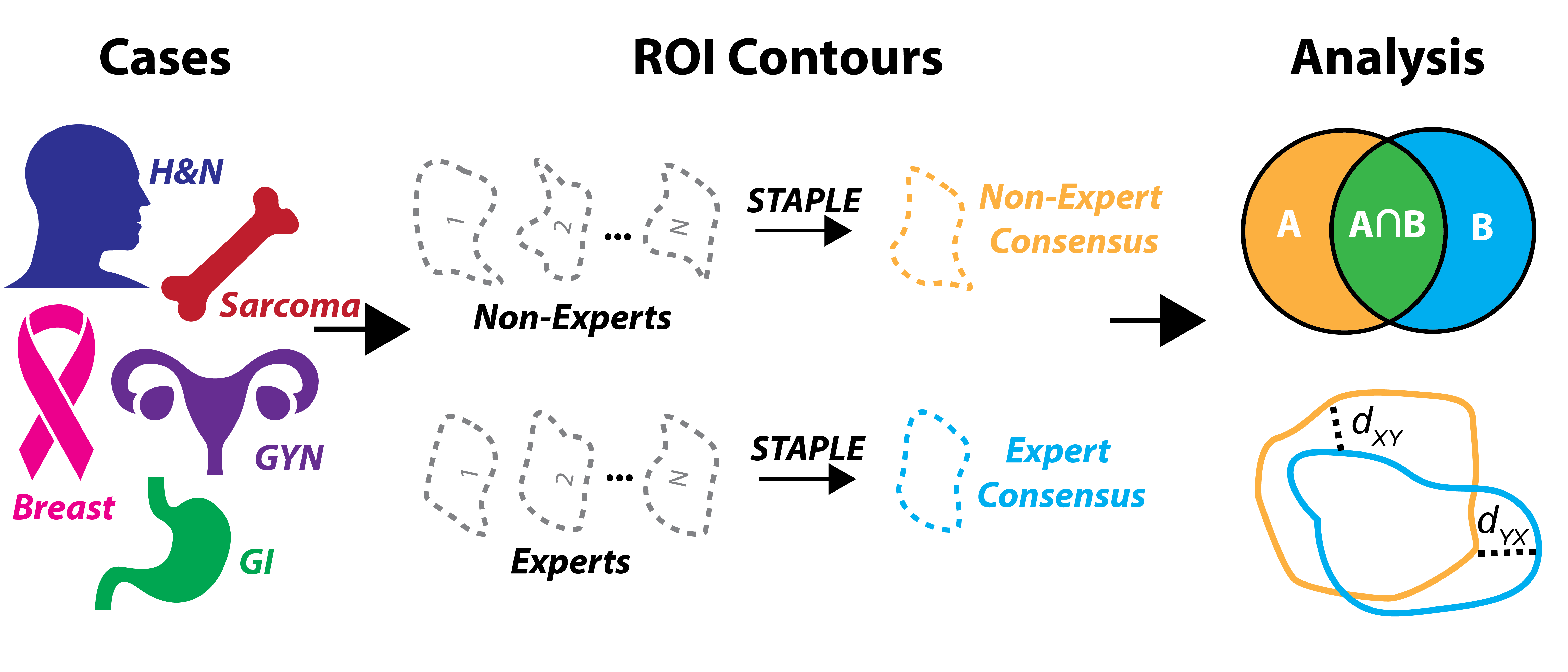
Prospective acceptability benchmarking from the Contouring Collaborative for Consensus in Radiation Oncology (C3RO) Crowdsourced Initiative for Multi-Observer Segmentation
Repo for code related to C3RO project. Manuscript available at: https://www.spiedigitallibrary.org/journals/journal-of-medical-imaging/volume-10/issue-S1/S11903/E-pluribus-unum--prospective-acceptability-benchmarking-from-the-Contouring/10.1117/1.JMI.10.S1.S11903.full?SSO=1. Corresponding image sets used for this project are avaliable on Figshare (data doi: 10.6084/m9.figshare.21074182). Data descriptor located at: https://www.nature.com/articles/s41597-023-02062-w. All data has been anonymized to remove patient PHI.
Jupyter notebook of main script (C3RO.ipynb) - This notebook contains all the code neccessary to perform the analyses outlined in the manuscript.
Pickle files associated with pairwise interobserver measurements (df_full_breast_allmetrics_expvsnonexp.pkl, df_full_sarcoma_final.pkl, df_full_H&N_allmetrics_expvsnonexp.pkl, df_full_GYN.pkl, df_full_GI.pkl) - Python Pickle files that were used as intermediates in generating results. Saved as intermediates because of long time to calculate.
Pickle files associated with bootstrap experiments (bootstrap100_expvsnonexp.pkl, bootstrap100_sarcoma.pkl, bootstrap100_H&N_expvsnonexp.pkl, bootstrap100_GYN.pkl, bootstrap100_GI.pkl) - Python Pickle files that were used to save bootstrap results. Saved as seperate files you can reference later because of long time to calculate 100 bootstraps.
DicomRTTool version 0.4.2.
SimpleITK version 2.1.1.
Numpy version 1.20.1.
Pandas version 1.4.2.
Surface-distance vresion 0.1
Nibabel version 4.0.1.
Statannotations version 0.4.4
Matplotlib version 3.5.1.
Seaborn version 0.11.2.