Supporting code for "Analysis of compounds activity concept learned by SVM using robust Jaccard based low-dimensional embedding" paper.
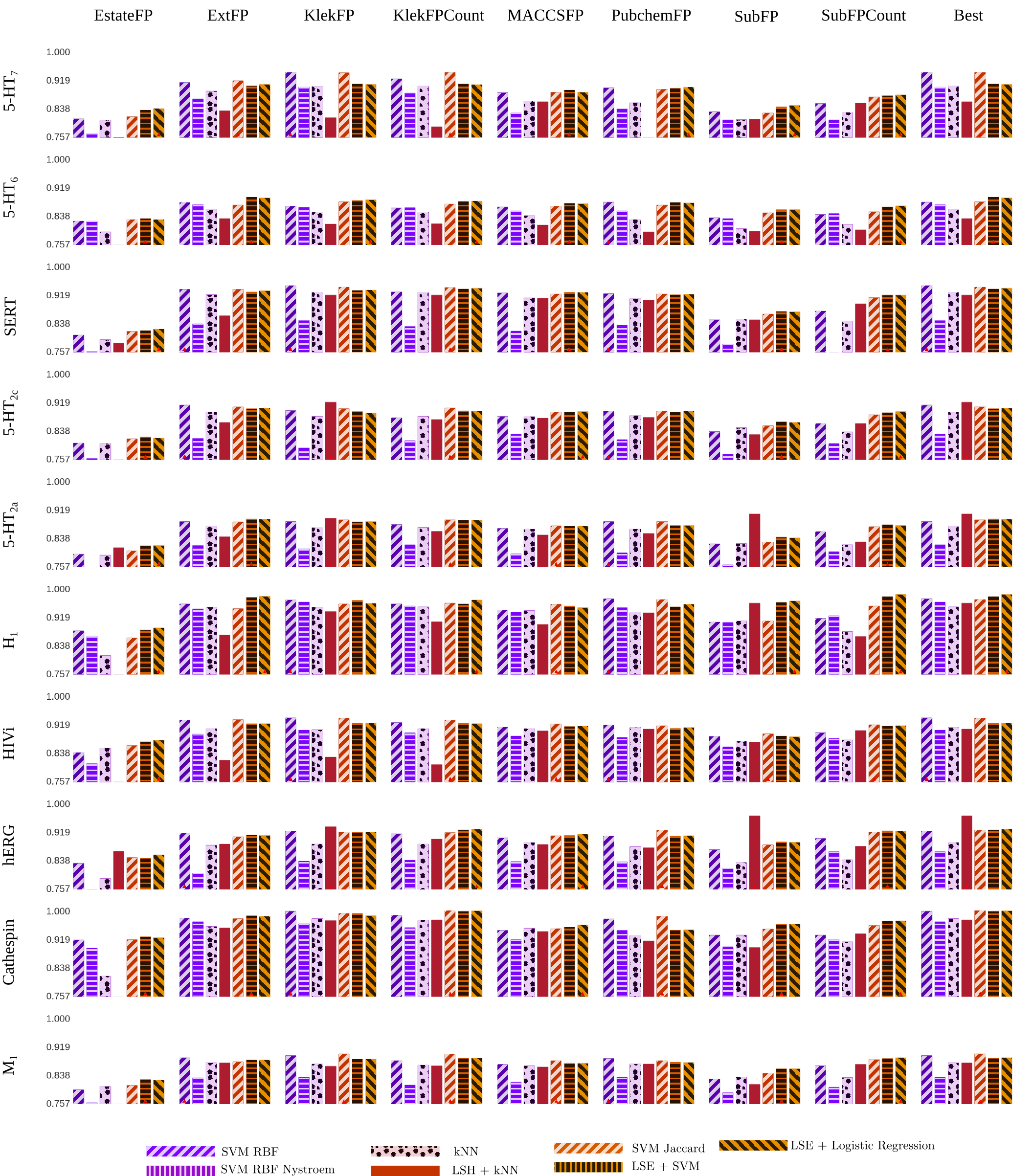
The paper deals with analysis of compound activity predictions. It argues that concepts learned by vastly popular SVM are degenerate given used sampling technique.
Change your misc/config.py file to match base directory and upload to data directory all .libsvm files
Scripts fit_knn.py, fit_svm.py, fit_lr.py, fit_melc.py are used to fit models. See python fit_<model>.py -h for usage.
Files in scripts folder were used to schedule many fittings, precalculate kernels for Jaccard etc. Each fitting script
writes experiment file that can be printed out.
To fit svms run python fit_svms.py.
Usage: fit_svms.py [options]
Options:
-h, --help show this help message and exit
-e EXPERIMENT_NAME, --e_name=EXPERIMENT_NAME
--kernel=KERNEL
--experiment_name=EXPERIMENT_NAME
--seed=SEED
--use_embedding=USE_EMBEDDING
--fingerprint=FINGERPRINT
--n_folds=N_FOLDS
--protein=PROTEIN
--max_hashes=MAX_HASHES
--grid_w=GRID_W
--K=K
Example
python fit_svms.py --kernel=linear --use_embedding=1 --protein=0 --fingerprint=4 --e_name=my_favourite_experiment
python scripts/fit_svms_print_results.py my_favourite_experiment