The Fiber Architecture Simulation Toolbox for 3D-PLI (fastpli) is a toolbox for polarized light imaging (PLI) with three main purposes:
-
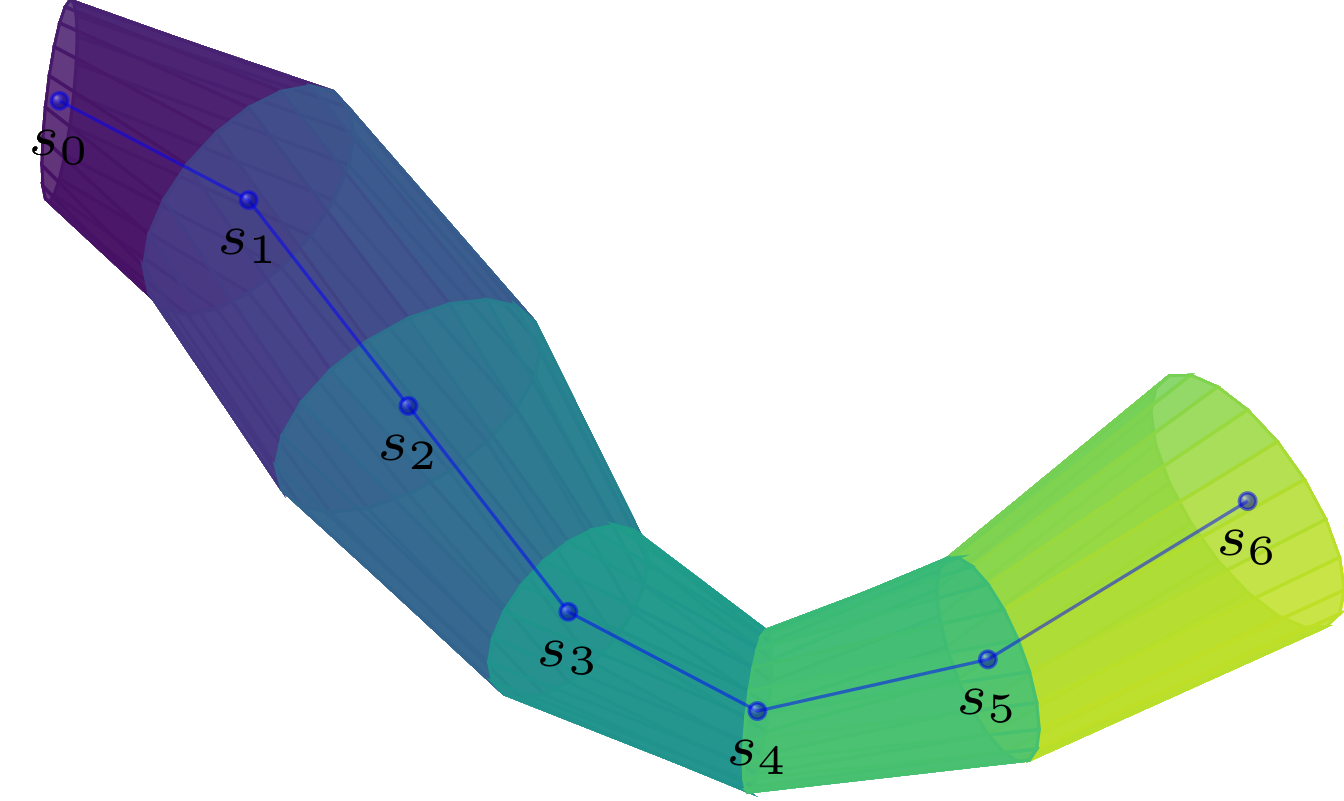
Sandbox- designing of nerve fiber models: The first module allows the user to create different types of nerve fiber bundles and additionally fill them with individual nerve fibers.
-
Solver- generating collision free models: The second module takes as input a configuration of nerve fibers and checks them for spatial collisions. Since nerve fibers cannot overlap in reality, one must ensure that the models follow the same rules. The solver module implements a simple algorithm that checks for collisions and, if it finds any, pushes the colliding segments of the fibers slightly apart. This is repeated until all collisions are solved.
-
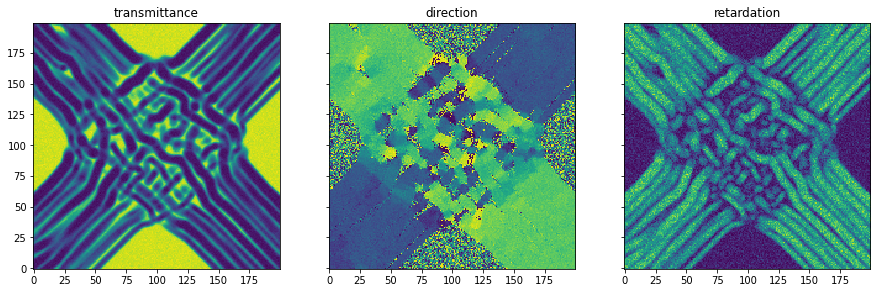
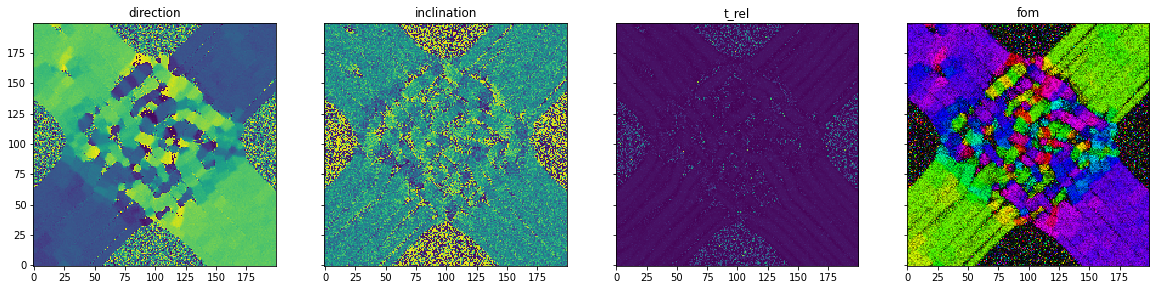
Simulation- simulation of 3D-Polarized Light Imaging: The simulation module enables the simulation of 3D Polarized Light Imaging (3D-PLI). This is a microscopic technique that allows the polarization change of light moving through a brain section to be measured. Due to the birefringence property of the myelin surrounding the nerve fibers, the polarization state changes. This change enables the calculation of the 3d orientation of the nerve fibers in the brain slice.
https://github.com/3d-pli/fastpli/wiki
As an example, a simplified model of the optic chiasm is presented. This structure in the brain allows nerve fibers from the eyes to cross each other and connect to the opposite side of the brain. In addition, a certain portion remains on the same side of the brain.
| module | information |
|---|---|
fastpli.analysis |
analysis of 3D-PLI results |
fastpli.io |
input/output functions, e.g. to read/save fiber_bundles data |
fastpli.model.sandbox |
building of simple 3d nerve fiber models |
fastpli.model.solver |
generation of non intersection nerve fiber models |
fastpli.objects |
manipulation of fastpli objects (e.g. rotation) |
fastpli.tools |
mathematical tools and helper function |
fastpli.simulation |
simulation of fiber models inside a virtual 3D-PLI microscope |
The current version of
fastplican only be run under Linux as operating system due to dependencies. If you want to usefastpliunder Windows, please use the Windows subsystem for Linux. To enable graphical output, you must install an X server. For more information, see https://wiki.ubuntu.com/WSL. Support for macOS is planned for the future.
- C++17
- Make
- CMake
- Python3
- MPI
- OpenGL (optional, recommended)
- pybind11
Install all necessary packages.
For Ubuntu:
sudo apt install gcc g++ cmake make git
sudo apt install python3-dev python3-venv
sudo apt install libopenmpi-dev freeglut3-devgit clone --recursive https://github.com/3d-pli/fastpli.git
cd fastpliUse your favorite environment e. g. python3 -m venv env and source env/bin/activate.
Probably you also have to update your pip version pip3 install pip -U.
make fastpli
pip3 install .# install required modules for examples
pip3 install -r examples/requirements.txt
jupyter-notebook examples/sandbox.ipynb
jupyter-notebook examples/solver.ipynb
jupyter-notebook examples/simulation.ipynb
jupyter-notebook examples/optic_chiasm.ipynb# install required modules for examples
pip3 install -r examples/requirements.txt
# run examples
python3 examples/sandbox.py
python3 examples/solver.py
python3 examples/simulation.py
python3 examples/optic_chiasm.pypython3 setup.py testAll computationally intensive calculations are optimized either with numba on the Python side or with multithreading C++, which can be accessed via pybind11. Additionally the simulation module supports the Message Passing Interface (MPI).
Please submit issues on GitHub to report
problems or suggest features. Pull requests
are also welcome to add features or correct problems.
Please run the local env-CI environment ./CI/run-all.sh or docker container make docker in advance.
- Felix Matuschke
fastPLI is an open source toolbox for modeling nerve fibers, simulating them in a 3D-PLI microscope and the signal processing developed by the fiber architecture group at the Forschungszentrum Jülich - INM1. This project has received funding from the European Union’s Horizon 2020 Research and Innovation Programme under Grant Agreement No. 7202070 (Human Brain Project SGA2).
 |
Forschungszentrum Jülich |
| Fiber Architecture - INM1 - Forschungszentrum Jülich | |
 |
Human Brain Project |
This project is licensed under the MIT License - see the LICENSE file for details