This is the official repository for the redevelopment of PConPy. The original
(now obsolete) source code associated with the article is accessible from the legacy branch of this repository.
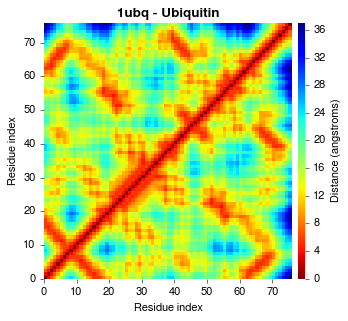
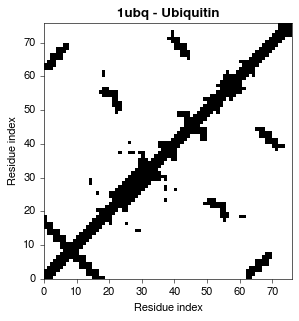
A contact map is a 2D representation of protein structure that illustrates the presence or absence of contacts between individual amino acids. This enables the rapid visual exploratory data analysis of structural features without 3D rendering software. Additionally, the underlying 2D matrix of a contact map, known as a contact matrix, can naturally be used as numeric input in subsequent automated knowledge discovery or machine learning tasks. PConPy generates publication-quality renderings of contact maps, and the related distance- and hydrogen bond maps.
Please consider citing our paper if you found PConPy to be useful in your research:
- H. K. Ho, M. Kuiper and K. Ramamohanarao, "PConPy—a Python module for generating 2D protein maps", Bioinformatics, vol. 24, no. 24, pp. 2934-2935, 2008. [article]
PConPy was developed using Python 2.7 using the following libraries:
- NumPy
- BioPython
- Matplotlib
- docopt
which can be installed via apt-get using Ubuntu:
sudo apt-get install python-numpy python-biopython python-matplotlib python-docopt
or via the Anaconda Python Distribution:
conda install numpy biopython matplotlib pip
pip install docopt
PConPy uses the DSSP secondary structure assignment program to obtain
inter-residue hydrogen bond information. The DSSP executable needs to be
installed into your system path and renamed to dssp, it can be
downloaded from:
- ftp://ftp.cmbi.ru.nl/pub/software/dssp/
Generate a PDF contact map using the CA-CA distance measure:
python ./pconpy/pconpy.py cmap 8.0 --pdb ./tests/pdb_files/1ubq.pdb \
--chains A --output 1ubqA_cmap.pdf --measure CA
Generate a PNG distance map using the min. VDW distance measure:
python ./pconpy/pconpy.py dmap --pdb ./tests/pdb_files/3erd.pdb \
--chains B,C --output 3erdBC_dmap.png --measure minvdw
Generate a plain-text hydrogen bond matrix:
python ./pconpy/pconpy.py hbmap --pdb ./tests/pdb_files/1ubq.pdb \
--chains A --plaintext --output 1ubq.txt
-
B. Konopka, M. Ciombor, M. Kurczynska and M. Kotulska, "Automated Procedure for Contact-Map-Based Protein Structure Reconstruction", The Journal of Membrane Biology, vol. 247, no. 5, pp. 409-420, 2014. [article]
-
A. Stivala, A. Wirth and P. Stuckey, Tableau-based protein substructure search using quadratic programming, BMC Bioinformatics, vol. 10, no. 1, p. 153, 2009. [article]
- Peter Cock's (@peterjc) tutorial covers the basics of PDB file parsing and visualisation using the powerful Biopython library.
See CONTRIBUTORS.md