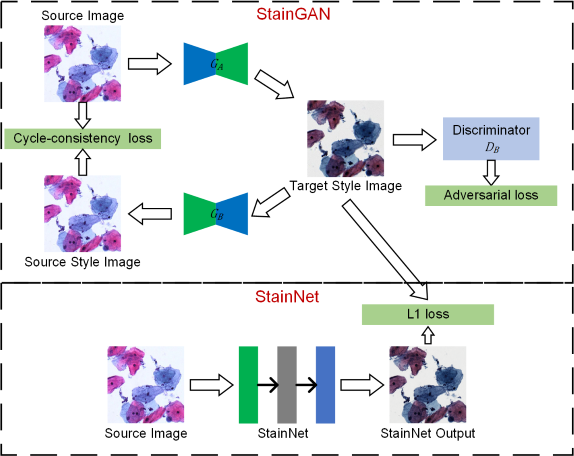
The code and dataset for StainNet with url: https://doi.org/10.3389/fmed.2021.746307 please cite our paper if you use the code or dataset
You can see the demo.ipynb for details
Python 3.6 or later with all requirements.txt dependencies installed, including torch>=1.0. To install run:
pip install -r requirements.txtWe make the aligned histopathology dataset and the histopathology classification dataset in our paper publicly available. They can be downloaded from BaiduYun as follow:
BaiduYun url:https://pan.baidu.com/s/1_k7l3wL0vrP26Yc6kkcWEQ Extraction code:wrfi
The related descriptions are below:
| Filename | Descriptions |
|---|---|
| aperio_hamamatsu.zip | The aligned histopathology dataset from the publicly available part of the MITOS-ATYPIA ICPR’14 challenge. |
| camelyon16.zip | The histopathology classification dataset from the publicly available Camelyon16 dataset for train classifier |
| camelyon16_for_train_norm.zip | From the publicly available Camelyon16 dataset for train normalization model |
git clone https://github.com/khtao/StainGAN.git
cd StainGAN
# train the aligned histopathology dataset
python train.py --dataroot aperio_hamamatsu/train --phaseA aperio --phaseB hamamatsu --batchSize 4 --niter 25 --niter_decay 25 --loadSize 256 --fineSize 256 --name aperio_hamamatsu --display_env aperio-hamamatsu --model cycle_gan --no_dropout
# train the histopathology classification dataset for norm
python train.py --dataroot camelyon16_for_train_norm --phaseA centerUni --phaseB centerRad --batchSize 4 --niter 100 --niter_decay 100 --loadSize 256 --fineSize 256 --name aperio_hamamatsu --display_env aperio-hamamatsu --model cycle_gan --no_dropout python test.py -h
--source_dir SOURCE_DIR
path to source images for test
--gt_dir GT_DIR path to ground truth images for test
--method METHOD different methods for test must be one of { StainNet StainGAN reinhard macenko vahadane khan }
--test_ssim whether calculate SSIM , default is False
--random_target random choose target or using matched ground truth, True is random choose target
--input_nc INPUT_NC # of input image channels
--output_nc OUTPUT_NC
# of output image channels
--channels CHANNELS # of channels in StainNet
--n_layer N_LAYER # of layers in StainNet
--model_path MODEL_PATH
models path to load
python train_StainNet.py -h
--name NAME name of the experiment.
--source_root_train SOURCE_ROOT_TRAIN
path to source images for training
--gt_root_train GT_ROOT_TRAIN
path to ground truth images for training
--source_root_test SOURCE_ROOT_TEST
path to source images for test
--gt_root_test GT_ROOT_TEST
path to ground truth images for test
--input_nc INPUT_NC # of input image channels
--output_nc OUTPUT_NC
# of output image channels
--channels CHANNELS # of channels in StainNet
--n_layer N_LAYER # of layers in StainNet
--batchSize BATCHSIZE
input batch size
--nThreads NTHREADS # threads for loading data
--checkpoints_dir CHECKPOINTS_DIR
models are saved here
--fineSize FINESIZE crop to this size
--display_freq DISPLAY_FREQ
frequency of showing training results on screen
--test_freq TEST_FREQ
frequency of cal
--lr LR initial learning rate for SGD
--epoch EPOCH how many epoch to train
python test_fps.py#train Classifier
python train_Classifier.py
#evel Classifier
python eval_Classifier.py