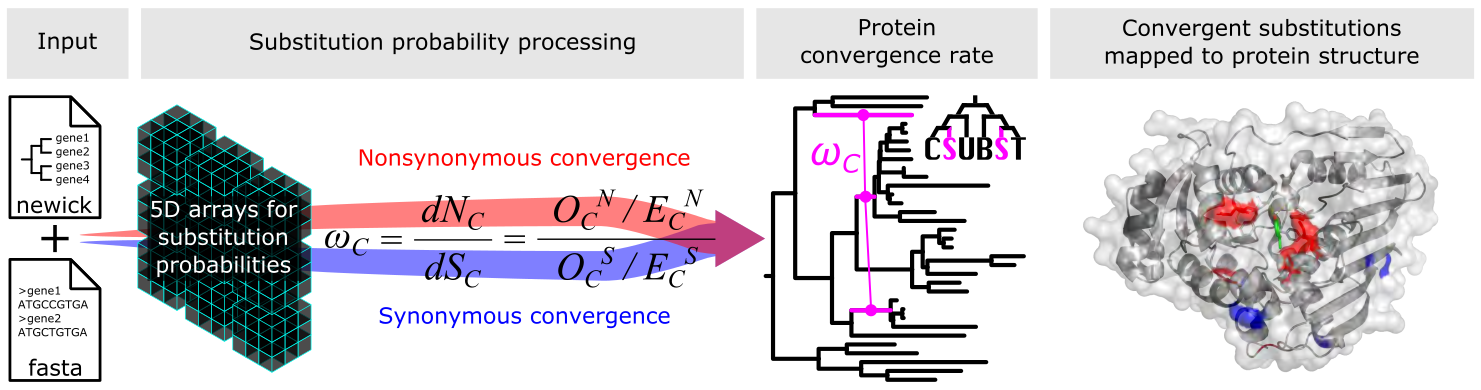
CSUBST (/si:sʌbst/) is a tool for analyzing Combinatorial SUBSTitutions of codon sequences in phylogenetic trees. A combinatorial substitution is defined as recurrent substitutions that occur at the same protein site in multiple independent branches. If multiple substitutions result in the same amino acid, they are considered convergent amino acid substitutions. The main features of CSUBST include:
- Error-corrected rate of protein convergence with null expectation obtained by:
- Empirical or mechanistic codon substitution model
- Urn sampling from site-wise substitution frequencies (experimental)
- Flexible specification of "foreground" lineages and its comparison with neighboring branches
- Heuristic detection of higher-order convergence involving more than two branches
- Simulated sequence evolution under specified scenarios of convergent evolution
- Convergent substitution mapping to protein structure
CSUBST takes as inputs:
- Newick file for the rooted tree
- FASTA file for the multiple sequence alignment of in-frame coding sequences
CSUBST runs on python 3 (tested with >=3.6.0). For a quick installation and test run, try:
# IQ-TREE installation with conda
conda install iqtree
# Installation with pip
pip install numpy cython # NumPy and Cython should be available upon csubst installation
pip install git+https://github.com/kfuku52/csubst
# Generate a test dataset
csubst dataset --name PGK
# Run csubst analyze
csubst analyze \
--alignment_file alignment.fa \
--rooted_tree_file tree.nwk \
--foreground foreground.txt
CSUBST is composed of several subcommands.
csubst -h shows the list of subcommands, and the complete set of subcommand options are available from csubst SUBCOMMAND -h (e.g., csubst analyze -h).
Many options are available, but those used by a typical user would be as follows.
More advanced usage is available in CSUBST wiki.
csubst datasetgenerates out-of-the-box test datasets.--name: Name of dataset. For a small test dataset, tryPGK(vertebrate phosphoglycerate kinase genes).
csubst analyzecalculates convergence rates and other metrics including ωC, dNC, and dSC on branch combinations.--alignment_file: PATH to input in-frame codon alignment.--rooted_tree_file: PATH to input rooted tree. Tip labels should be consistent with--alignment_file.--genetic_code: NCBI codon table ID. 1 = "Standard". See here for details.--iqtree_model: Codon substitution model for ancestral state reconstruction. Base models of "MG", "GY", "ECMK07", and "ECMrest" are supported. Among-site rate heterogeneity and codon frequencies can be specified. See IQTREE's website for details.--threads: The number of CPUs for parallel computations (e.g.,1or4).--foreground: Optional. A text file to specify the foreground lineages. The file should contain two columns separated by a tab: 1st column for lineage IDs and 2nd for regex-compatible leaf names.
csubst sitecalculates site-wise combinatorial substitutions on focal branch combinations and maps it onto protein structure.--alignment_file: PATH to input in-frame codon alignment.--rooted_tree_file: PATH to input rooted tree. Tip labels should be consistent with--alignment_file.--genetic_code: NCBI codon table ID. 1 = "Standard". See here for details.--iqtree_model: Codon substitution model for ancestral state reconstruction. Base models of "MG", "GY", "ECMK07", and "ECMrest" are supported. Among-site rate heterogeneity and codon frequencies can be specified. See IQTREE's website for details.
csubst simulategenerates a simulated sequence alignment under a convergent evolutionary scenario.--alignment_file: PATH to input in-frame codon alignment.--rooted_tree_file: PATH to input rooted tree. Tip labels should be consistent with--alignment_file.--genetic_code: NCBI codon table ID. 1 = "Standard". See here for details.--iqtree_model: Codon substitution model for ancestral state reconstruction. Base models of "MG", "GY", "ECMK07", and "ECMrest" are supported. Among-site rate heterogeneity and codon frequencies can be specified. See IQTREE's website for details.--foreground: A text file to specify the foreground lineages. The file should contain two columns separated by a tab: 1st column for lineage IDs and 2nd for regex-compatible leaf names.
Fukushima K, Pollock DD. 2023. Detecting macroevolutionary genotype-phenotype associations using error-corrected rates of protein convergence. Nature Ecology & Evolution 7: 155–170. DOI: 10.1038/s41559-022-01932-7
CSUBST is BSD-licensed (3 clause). See LICENSE for details.