| Release | Usage | Development |
|---|---|---|
 |
||
 |
||
The drake R package 
The drake R package is a workflow manager and computational engine for data science projects. Its primary objective is to keep results up to date with the underlying code and data. When it runs a project, drake detects any pre-existing output and refreshes the pieces that are outdated or missing. Not every runthrough starts from scratch, and the final answers are reproducible. With a user-friendly R-focused interface, comprehensive documentation, and extensive implicit parallel computing support, drake surpasses the analogous functionality in similar tools such as Make, remake, memoise, and knitr.
What gets done stays done.
Too many data science projects follow a Sisyphean loop:
- Launch the code.
- Wait for it to finish.
- Discover an issue.
- Restart from scratch.
But drake automatically
- Launches the parts that changed since the last runthrough.
- Skips the rest.
library(drake)
# Drake comes with a basic example.
# Get the code with drake_example("basic").
load_basic_example(verbose = FALSE)
# Your workspace starts with a bunch of "imports":
# functions, pre-loaded data objects, and saved files
# available before the real work begins.
# Drake looks for data objects in your R session environment
ls()
## [1] "my_plan" "random_rows" "reg1" "reg2" "simulate"
# and saved files in your file system.
list.files()
## [1] "report.Rmd"
# The real work is outlined step-by-step in the `my_plan` data frame.
# The steps are called "targets", and they depend on the imports.
# File targets have the names in `file_out()`, and the non-file
# targets have the names in the `target` column of the data frame.
# Drake's `make()` function runs the commands to build the targets
# in the correct order.
head(my_plan)
## # A tibble: 6 x 2
## target command
## <chr> <chr>
## 1 "" "knit(knitr_in(\"report.Rmd\"), file_out(\"report.md\"), quiet = TRUE)"
## 2 small simulate(48)
## 3 large simulate(64)
## 4 regression1_small reg1(small)
## 5 regression1_large reg1(large)
## 6 regression2_small reg2(small)
# First round: drake builds all 15 targets.
make(my_plan)
## target large
## target small
## target regression1_large
## target regression1_small
## target regression2_large
## target regression2_small
## target coef_regression1_large
## target coef_regression1_small
## target coef_regression2_large
## target coef_regression2_small
## target summ_regression1_large
## target summ_regression1_small
## target summ_regression2_large
## target summ_regression2_small
## target "report.md"
# If you change the reg2() function,
# all the regression2 targets are out of date,
# which in turn affects 'report.md'.
reg2 <- function(d){
d$x4 <- d$x ^ 4
lm(y ~ x4, data = d)
}
# Second round: drake only rebuilds the targets
# that depend on the things you changed.
make(my_plan)
## target regression2_large
## target regression2_small
## target coef_regression2_large
## target coef_regression2_small
## target summ_regression2_large
## target summ_regression2_small
## target "report.md"
# If nothing important changed, drake rebuilds nothing.
make(my_plan)
## All targets are already up to date.
# Drake cares about nested functions too:
# with the exception of trivial formatting edits,
# changes to `random_rows()` will propagate to `simulate()`
# and all the downstream targets.
# Try it!Reproducibility with confidence
The R community emphasizes reproducibility. Traditional themes include scientific replicability, literate programming with knitr, and version control with git. But internal consistency is important too. Reproducibility carries the promise that your output matches the code and data you say you used.
Evidence
Suppose you are reviewing someone else's data analysis project for reproducibility. You scrutinize it carefully, checking that the datasets are available and the documentation is thorough. But could you re-create the results without the help of the original author? With drake, it is quick and easy to find out.
make(my_plan)
## All targets are already up to date.
config <- drake_config(my_plan)
outdated(config)
## character(0)With everything already up to date, you have tangible evidence of reproducibility. Even though you did not re-create the results, you know the results are re-creatable. They faithfully show what the code is producing. Given the right package environment and system configuration, you have everything you need to reproduce all the output by yourself.
Ease
When it comes time to actually rerun the entire project, you have much more confidence. Starting over from scratch is trivially easy.
clean() # Remove the original author's results.
make(my_plan) # Independently re-create the results from the code and input data.Independent replication
With even more evidence and confidence, you can invest the time to independently replicate the original code base if necessary. Up until this point, you relied on basic drake functions such as make(), so you may not have needed to peek at any substantive author-defined code in advance. In that case, you can stay usefully ignorant as you reimplement the original author's methodology. In other words, drake could potentially improve the integrity of independent replication.
Readability and transparency
For reproducibility, it is important that others can read your code and understand what your workflow is doing. Drake helps in several ways.
- The workflow plan data frame explicitly outlines the steps of the analysis, and
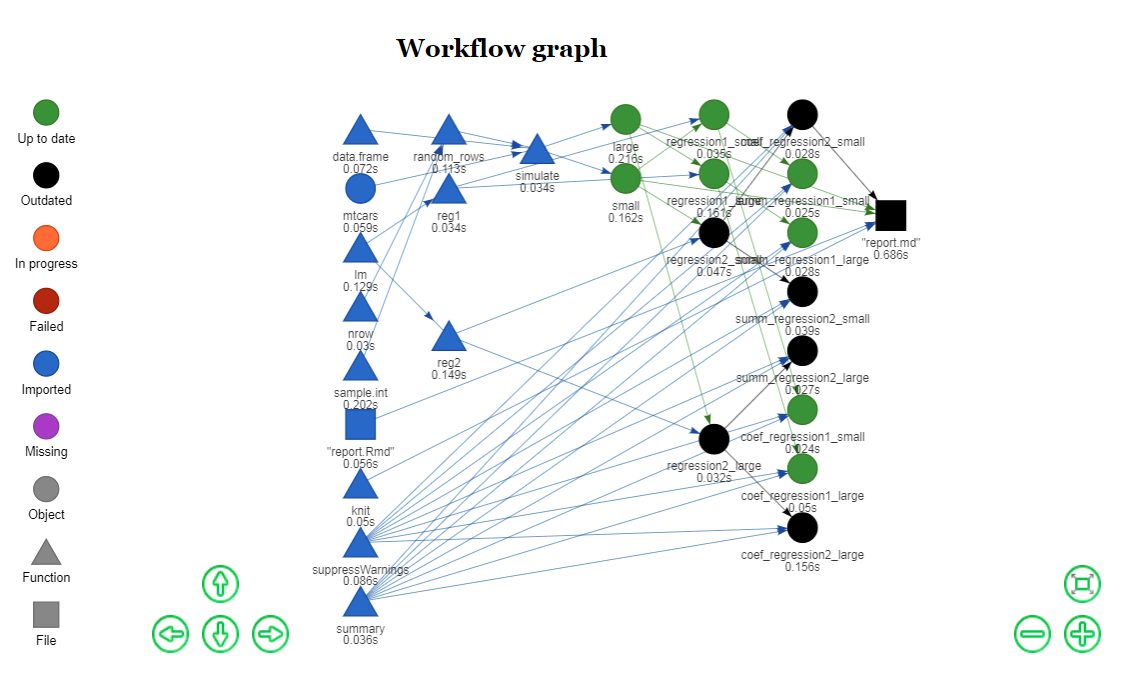
vis_drake_graph()visualizes how those steps depend on each other. Draketakes care of the parallel scheduling and high-performance computing (HPC) so you do not have to. That means the HPC code is no longer tangled up with the code that actually expresses your ideas.- You can generate large numbers of targets without encumbering the collection of imported functions that make up your code base. In other words,
drakecan scale up the size of your project without making your custom code more complicated.
Aggressively scale up.
Not every project can complete in a single R session on your laptop. Some projects need more speed or computing power. Some require a few local processor cores, and some need large high-performance computing systems. But parallel computing is hard. Your tables and figures depend on your analysis results, and your analyses depend on your datasets, so some tasks must finish before others even begin. But drake knows what to do. Parallelism is implicit and automatic. See the parallelism vignette for all the details.
# Use the spare cores on your local machine.
make(my_plan, jobs = 4)
# Scale up to a supercomputer.
drake_batchtools_tmpl_file("slurm") # https://slurm.schedmd.com/
library(future.batchtools)
future::plan(batchtools_slurm, template = "batchtools.slurm.tmpl", workers = 100)
make(my_plan, parallelism = "future_lapply")The network graph allows drake to wait for dependencies.
# Change some code.
reg2 <- function(d){
d$x3 <- d$x ^ 3
lm(y ~ x3, data = d)
}
# Plot an interactive graph.
config <- drake_config(my_plan)
vis_drake_graph(config)Within each column above, the nodes are conditionally independent given their dependencies. Each make() walks through the columns from left to right and applies parallel processing within each column. If any nodes are already up to date, drake looks downstream to maximize the number of outdated targets in a parallelizable stage. To show the parallelizable stages of the next make() programmatically, use the parallel_stages() function.
Installation
You can choose among different versions of drake.
# Install the latest stable release from CRAN.
install.packages("drake")
# Alternatively, install the development version from GitHub.
install.packages("devtools")
library(devtools)
install_github("ropensci/drake")- You must properly install
drakeusinginstall.packages(),devtools::install_github(), or similar. It is not enough to usedevtools::load_all(), particularly for the parallel computing functionality, in which multiple R sessions initialize and then try torequire(drake). - For
make(..., parallelism = "Makefile"), Windows users need to download and installRtools. - If you want to build the vignettes when you install the development version, you must
- Install all the packages in the
Suggests:field of the DESCRIPTION file, including cranlogs and Ecdat. All these packages are available through the Comprehensive R Archive Network (CRAN), and you can install them withinstall.packages(). - Set the
buildargument toTRUEininstall_github().
- Install all the packages in the
Documentation
The main resources to learn drake are
Cheat sheet
Thanks to Kirill for preparing a drake cheat sheet for the workshop.
Frequently asked questions
The FAQ page is an index of links to appropriately-labeled issues on GitHub. To contribute, please submit a new issue and ask that it be labeled as a frequently asked question.
Function reference
The reference section lists all the available functions. Here are the most important ones.
drake_plan(): create a workflow data frame (likemy_plan).make(): build your project.loadd(): load one or more built targets into your R session.readd(): read and return a built target.drake_config(): create a master configuration list for other user-side functions.vis_drake_graph(): show an interactive visual network representation of your workflow.outdated(): see which targets will be built in the nextmake().deps(): check the dependencies of a command or function.failed(): list the targets that failed to build in the lastmake().diagnose(): return the full context of a build, including errors, warnings, and messages.
Tutorials
Thanks to Kirill for constructing two interactive learnr tutorials: one supporting drake itself, and a prerequisite walkthrough of the cooking package.
The articles below are tutorials taken from the package vignettes.
- Get started
- Example: R package download trends
- Example: gross state products
- Basic example
- General best practices
- Cautionary notes and edge cases
- Debugging and testing drake projects
- Graphs with drake
- Parallel computing
- Time logging
- Storage
Examples
Drake also has built-in example projects with code files available here. You can generate the files for a project with drake_example() (e.g. drake_example("gsp")), and you can list the available projects with drake_examples(). The beginner-oriented examples are listed below. They help you learn drake's main features, and they show one way to organize the files of drake projects.
basic: A tiny, minimal example with themtcarsdataset to demonstrate how to usedrake. Useload_basic_example()to set up the project in your workspace. The quickstart vignette is a parallel walkthrough of the same example.gsp: A concrete example using real econometrics data. It explores the relationships between gross state product and other quantities, and it shows offdrake's ability to generate lots of reproducibly-tracked tasks with ease.packages: A concrete example using data on R package downloads. It demonstrates howdrakecan refresh a project based on new incoming data without restarting everything from scratch.
Presentations
- https://krlmlr.github.io/drake-pitch
- https://krlmlr.github.io/slides/drake-sib-zurich
- https://krlmlr.github.io/slides/drake-sib-zurich/cooking.html
Context and history
For context and history, check out this post on the rOpenSci blog and episode 22 of the R Podcast.
Help and troubleshooting
The following resources document many known issues and challenges.
- Frequently-asked questions.
- Cautionary notes and edge cases
- Debugging and testing drake projects
- Other known issues (please search both open and closed ones).
If you are still having trouble, please submit a new issue with a bug report or feature request, along with a minimal reproducible example where appropriate.
Contributing
Bug reports, suggestions, and especially code contributions are welcome. Please see CONTRIBUTING.md. Maintainers and contributors must follow this repository's code of conduct.
Similar work
GNU Make
The original idea of a time-saving reproducible build system extends back at least as far as GNU Make, which still aids the work of data scientists as well as the original user base of complied language programmers. In fact, the name "drake" stands for "Data Frames in R for Make". Make is used widely in reproducible research. Below are some examples from Karl Broman's website.
- Bostock, Mike (2013). "A map of flowlines from NHDPlus." https://github.com/mbostock/us-rivers. Powered by the Makefile at https://github.com/mbostock/us-rivers/blob/master/Makefile.
- Broman, Karl W (2012). "Halotype Probabilities in Advanced Intercross Populations." G3 2(2), 199-202.Powered by the
Makefileat https://github.com/kbroman/ailProbPaper/blob/master/Makefile. - Broman, Karl W (2012). "Genotype Probabilities at Intermediate Generations in the Construction of Recombinant Inbred Lines." *Genetics 190(2), 403-412. Powered by the Makefile at https://github.com/kbroman/preCCProbPaper/blob/master/Makefile.
- Broman, Karl W and Kim, Sungjin and Sen, Saunak and Ane, Cecile and Payseur, Bret A (2012). "Mapping Quantitative Trait Loci onto a Phylogenetic Tree." Genetics 192(2), 267-279. Powered by the
Makefileat https://github.com/kbroman/phyloQTLpaper/blob/master/Makefile.
There are several reasons for R users to prefer drake instead.
Drakealready has a Make-powered parallel backend. Just runmake(..., parallelism = "Makefile", jobs = 2)to enjoy most of the original benefits of Make itself.- Improved scalability. With Make, you must write a potentially large and cumbersome Makefile by hand. But with
drake, you can use wildcard templating to automatically generate massive collections of targets with minimal code. - Lower overhead for light-weight tasks. For each Make target that uses R, a brand new R session must spawn. For projects with thousands of small targets, that means more time may be spent loading R sessions than doing the actual work. With
make(..., parallelism = "mclapply, jobs = 4"),drakelaunches 4 persistent workers up front and efficiently processes the targets in R. - Convenient organization of output. With Make, the user must save each target as a file.
Drakesaves all the results for you automatically in a storr cache so you do not have to micromanage the results.
Remake
Drake overlaps with its direct predecessor, remake. In fact, drake owes its core ideas to remake and Rich Fitzjohn. Remake's development repository lists several real-world applications. Drake surpasses remake in several important ways, including but not limited to the following.
- High-performance computing. Remake has no native parallel computing support. Drake, on the other hand, has a vast arsenal of parallel computing options, from local multicore computing to serious distributed computing. Thanks to future, future.batchtools, and batchtools, it is straightforward to configure a drake project for most popular job schedulers, such as SLURM, TORQUE, and the Sun/Univa Grid Engine, as well as systems contained in Docker images.
- A friendly interface. In remake, the user must manually write a YAML configuration file to arrange the steps of a workflow, which leads to some of the same scalability problems as Make. Drake's data-frame-based interface and wildcard templating functionality easily generate workflows at scale.
- Thorough documentation. Drake contains several vignettes, a comprehensive README, examples in the help files of user-side functions, and accessible example code that users can write with
drake::example_drake(). - Active maintenance. Drake is actively developed and maintained, and issues are usually solved promptly.
- Presence on CRAN. At the time of writing, drake is available on CRAN, but remake is not.
Memoise
Memoization is the strategic caching of the return values of functions. Every time a memoized function is called with a new set of arguments, the return value is saved for future use. Later, whenever the same function is called with the same arguments, the previous return value is salvaged, and the function call is skipped to save time. The memoise package is an excellent implementation of memoization in R.
However, memoization does not go far enough. In reality, the return value of a function depends not only on the function body and the arguments, but also on any nested functions and global variables, the dependencies of those dependencies, and so on upstream. Drake surpasses memoise because it uses the entire dependency network graph of a project to decide which pieces need to be rebuilt and which ones can be skipped.
Knitr
Much of the R community uses knitr for reproducible research. The idea is to intersperse code chunks in an R Markdown or *.Rnw file and then generate a dynamic report that weaves together code, output, and prose. Knitr is not designed to be a serious pipeline toolkit, and it should not be the primary computational engine for medium to large data analysis projects.
- Knitr scales far worse than Make or remake. The whole point is to consolidate output and prose, so it deliberately lacks the essential modularity.
- There is no obvious high-performance computing support.
- While there is a way to skip chunks that are already up to date (with code chunk options
cacheandautodep), this functionality is not the focus of knitr. It is deactivated by default, and remake anddrakeare more dependable ways to skip work that is already up to date.
As in the basic example demonstrates, drake should manage the entire workflow, and any knitr reports should quickly build as targets at the very end. The strategy is analogous for knitr reports within remake projects.
Factual's Drake
Factual's Drake is similar in concept, but the development effort is completely unrelated to the drake R package.
Other pipeline toolkits
There are countless other successful pipeline toolkits. The drake package distinguishes itself with its R-focused approach, Tidyverse-friendly interface, and wide selection of parallel computing backends.
Acknowledgements
Special thanks to Jarad Niemi, my advisor from graduate school, for first introducing me to the idea of Makefiles for research. He originally set me down the path that led to drake.
Many thanks to Julia Lowndes, Ben Marwick, and Peter Slaughter for reviewing drake for rOpenSci, and to Maëlle Salmon for such active involvement as the editor. Thanks also to the following people for contributing early in development.
- Alex Axthelm
- Chan-Yub Park
- Daniel Falster
- Eric Nantz
- Henrik Bengtsson
- Ian Watson
- Jasper Clarkberg
- Kendon Bell
- Kirill Müller
Credit for images is attributed here.