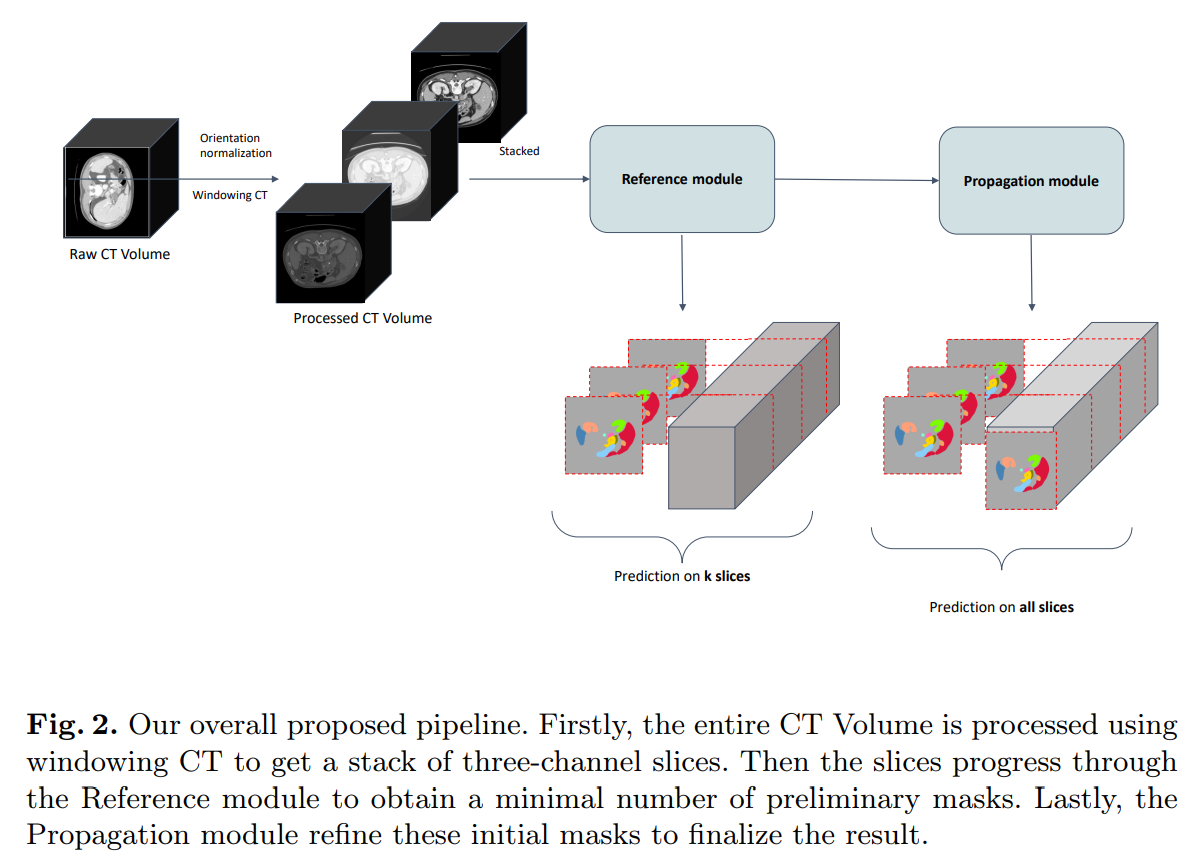
Semi-supervised organ segmentation with Mask Propagation Refinement and Uncertainty Estimation for Data Generation
We present a novel two-staged method that employs various 2D-based techniques to deal with the 3D segmentation task. In most of the previous challenges, it is unlikely for 2D CNNs to be comparable with other 3D CNNs since 2D models can hardly capture temporal information. In light of that, we propose using the recent state-of-the-art technique in video object segmentation, combining it with other semi-supervised training techniques to leverage the extensive unlabeled data. Moreover, we introduce a way to generate pseudo-labeled data that is both plausible and consistent for further retraining by using uncertainty estimation.
We also deploy an annotation tool with user-friendly GUI that is based on this proposed algorithm here
Our work has been published as part of the Lecture Notes in Computer Science book series (LNCS, volume 13816)
| Windows/Ubuntu version | Ubuntu 18.04.5 LTS |
|---|---|
| CPU | Intel(R) Xeon(R) Silver 4210R CPU @ 2.40GHz |
| RAM | 1×32GB; |
| GPU (number and type) | One Quadro RTX 5000 16G |
| CUDA version | 11.6 |
| Programming language | Python 3.10 |
| Deep learning framework | Pytorch (Torch 1.11.0, torchvision 0.12.0) |
- To reproduce the results that were reported in FLARE22 Challenge, please see Docker
- We reuploaded the original data from FLARE22 Challenge onto Drive, use below scripts to download and preprocess the data before training.
sh tools/scripts/download_data.sh
sh tools/scripts/preprocess_data.sh
- After processing, the data structure should be as followed:
this repo
│
└───data
| └───flare22
| └───raw
| └───slices # For training
| └───npy # For inference/evaluation
- In each of these folder should be subfolders for training/val/validation images and masks.
-
We use Cross Pseudo Supervision (CPS) for the Reference module. Basically, CPS can be trained from scratch but we recommend training each model individually first.
-
To train individual model, customize
configs/flare22v2/normal/pipeline.yamlthen run
sh tools/scripts/train_normal.sh <run_name> <save_dir>
- To train cps model, customize
configs/flare22v2/cps/pipeline.yamlthen run
sh tools/scripts/train_cps.sh <run_name> <save_dir>
-
We adapt STCN as the core algorithm to our problem with adjustments.
-
To train stcn model, customize
configs/flare22v2/stcn/pipeline.yamlthen run
sh tools/scripts/train_cps.sh <run_name> <save_dir>
- You can download the checkpoints from wandb by using
sh tools/scripts/download_weights.sh
which saves the trained weights to weights folder. These can be used to reproduce the results that we reported in the paper
- To perform 2-stage inference, please modify
configs/flare22v2/stcn/test.yaml, then run
sh tools/scripts/infer_stcn.sh
which will then save the results inside runs/<model_name>/. Submission file along with visualization will be saved here.
- For other inference settings, some scripts are also provided.
- We use Uncertainty Estimation based on mutual agreement to generate pseudo-labels for retraining. Put all results that are infered using provided inference scripts into the same <prediction_dir> folder. Then run
sh tools/scripts/pseudo_label.sh <prediction_dir> <save_dir>
- Standalone evaluation scripts officially provided by the organizers
python tools/evaluation/DSC_NSD_eval.py \
-g <ground truth mask> \
-p <prediction mask>
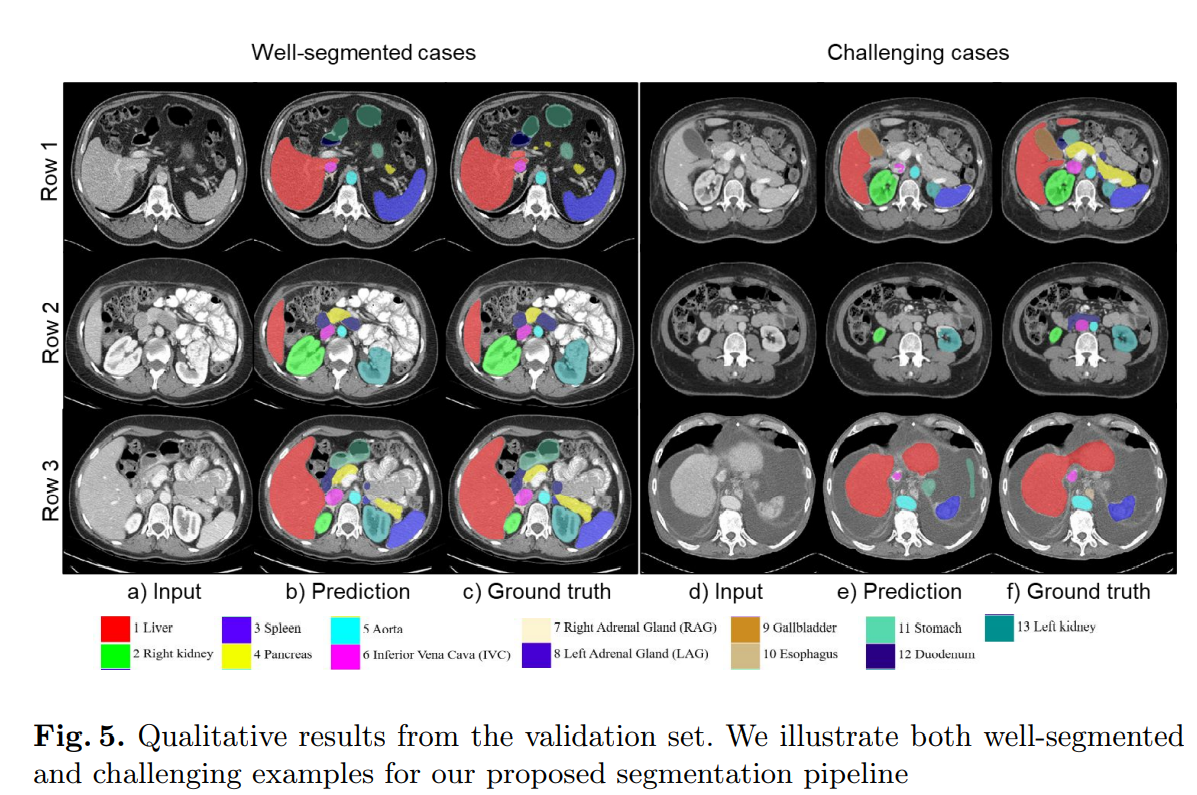
- The results are officially reported by the organizers of FLARE22 Challenge
| Classes/Metrics | DSC | NSD |
|---|---|---|
| Liver | 0.974 ± 0.036 | 0.963 ± 0.063 |
| Right Kidney (RK) | 0.883 ± 0.233 | 0.868 ± 0.241 |
| Spleen | 0.9494 ± 0.115 | 0.935 ± 0.134 |
| Pancreas | 0.772 ± 0.147 | 0.877 ± 0.145 |
| Aorta | 0.96 ± 0.045 | 0.976 ± 0.06 |
| Inferior Vena Cava (IVC) | 0.86 ± 0.123 | 0.86 ± 0.143 |
| Right Adrenal Gland (RAG) | 0.735 ± 0.138 | 0.855 ± 0.144 |
| Left Adrenal Gland (LAG) | 0.69 ± 0.171 | 0.816 ± 0.2 |
| Gallbladder | 0.75 ± 0.313 | 0.733 ± 0.328 |
| Esophagus | 0.783 ± 0.147 | 0.88 ± 0.143 |
| Stomach | 0.86 ± 0.113 | 0.84 ± 0.142 |
| Duodenum | 0.6 ± 0.2 | 0.79 ± 0.215 |
| Left Kidney (LK) | 0.877 ± 0.22 | 0.863 ± 0.23 |
| Mean | 0.8233 | 0.8668 |
- https://github.com/kaylode/theseus
- https://github.com/hkchengrex/STCN
- https://github.com/hkchengrex/MiVOS
- https://github.com/hkchengrex/XMem
@inproceedings{cheng2021stcn,
title={Rethinking Space-Time Networks with Improved Memory Coverage for Efficient Video Object Segmentation},
author={Cheng, Ho Kei and Tai, Yu-Wing and Tang, Chi-Keung},
booktitle={NeurIPS},
year={2021}
}
@inproceedings{cheng2021mivos,
title={Modular Interactive Video Object Segmentation: Interaction-to-Mask, Propagation and Difference-Aware Fusion},
author={Cheng, Ho Kei and Tai, Yu-Wing and Tang, Chi-Keung},
booktitle={CVPR},
year={2021}
}
@inproceedings{cheng2022xmem,
title={{XMem}: Long-Term Video Object Segmentation with an Atkinson-Shiffrin Memory Model},
author={Cheng, Ho Kei and Alexander G. Schwing},
booktitle={ECCV},
year={2022}
}