In Proceedings of the 31th ACM SIGKDD Conference on Knowledge Discovery and Data Mining (KDD '25), August 3-7, 2025, Toronto, ON, Canada.
Abstract: Proteins play a vital role in biological processes and are indispensable for living organisms. Accurate representation of proteins is crucial, especially in drug development. Recently, there has been a notable increase in interest in utilizing machine learning and deep learning techniques for unsupervised learning of protein representations. However, these approaches often focus solely on the amino acid sequence of proteins and lack factual knowledge about proteins and their interactions, thus limiting their performance. In this study, we present GOProteinGNN, a novel architecture that enhances language models by integrating protein knowledge graph information during the creation of amino acid-level representations. Our approach allows for the integration of information at both the individual amino acid level and the entire protein level, enabling a comprehensive and effective learning process through graph-based learning. By doing so, we can capture complex relationships and dependencies between proteins and their functional annotations, resulting in more robust and contextually enriched protein representations. Furthermore, GOProteinGNN sets itself apart by uniquely learning the entire protein knowledge graph during training which allows it to capture broader relational nuances and dependencies beyond mere triplets as done in previous works. We perform comprehensive evaluations on several downstream tasks demonstrating that GOProteinGNN consistently outperforms previous methods, showcasing its effectiveness and establishing it as a state-of-the-art solution for protein representation learning.
The GOProteinGNN pre-training architecture:
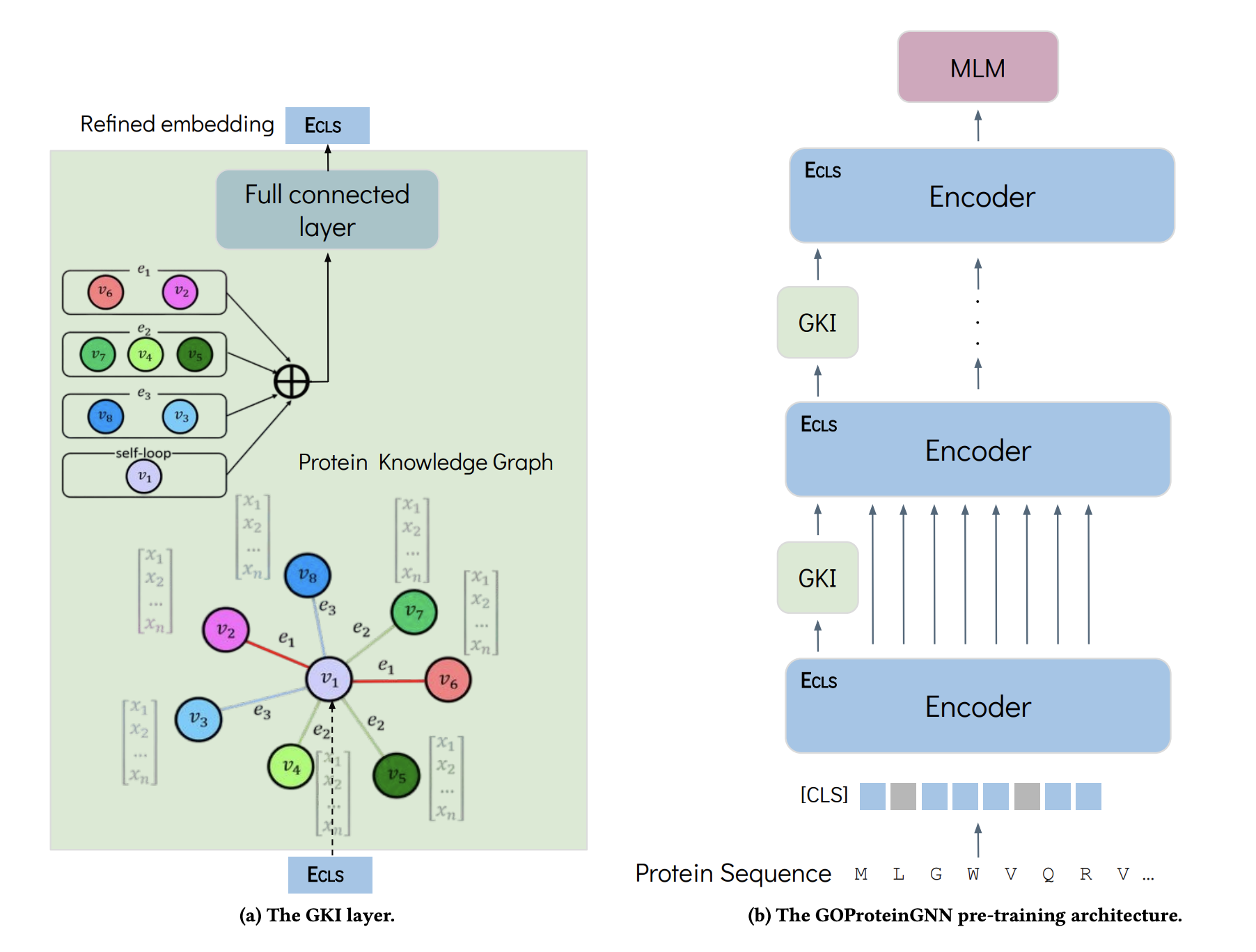
This repository provides a reference implementation of GOProteinGNN and baselines as described in the paper. Some code was borrowed from KeAP and OntoProtein.
ProteinKG25 is a large-scale knowledge graph dataset with aligned descriptions and protein sequences respectively to GO terms and protein entities. This dataset is necessary for performing pre-training. You can follow the instruction to configure ProteinKG25.
Main dependencies is listed in the requirments.txt file and build with Python 3.7. Execute the following command to install all required dependencies:
pip install -r requirements.txtFollowing OntoProtein, we also make small changes to the deepspeed.py file under transformers library (❗required for pre-training).
The changes can be applied by running:
cp replace_code/deepspeed.py <path-to-python3.7>/site-packages/transformers/deepspeed.pyMain dependencies is listed in the requirments.txt file and build with Python 3.7. Execute the following command to install all required dependencies:
pip install -r requirements.txtNote PyTorch Geometric is required for the PPI (protein-protein interaction) task. Check your PyTorch and cuda versions, and follow the installation instructions to install correctly.
Since the tape_proteins library only implemented the P@L metric for the contact prediction task, we add the P@L/5 and P@L/2 metrics by running the following script:
cp replace_code/tape/modeling_utils.py <path-to-python3.7>/site-packages/tape/models/modeling_utils.pyFor pre-training data preparation, please refer to here.
The data for TAPE tasks and the PPI task can be downloaded from here. The data for the PROBE tasks can be acquired via link.
After configuring ProteinKG25, you also need to download the following two pre-trained models:
- ProtBERT for initializing the protein encoder.
- PubMedBERT for extracting text features in Gene Onotology Annotations.
Then, configure paths in script/run_pretrain.sh (PRETRAIN_DATA_DIR, ENCODER_MODEL_PATH, TEXT_MODEL_PATH) accordingly.
Run the following script for pre-training:
sh ./script/run_pretrain.shThe detailed arguments are listed in src/training_args.py.
In this part, we fine-tune the pre-trained model (GOProteinGNN) on various downstream tasks.
❗NOTE: You will need to change some paths for downstream data and extracted embeddings (PPI and PROBE tasks) before running the code.
Secondary structure prediction, contact prediction, remote homology detection, stability prediction, and fluorescence prediction are tasks from TAPE.
Similar to KeAP and OntoProtein, for these tasks, we provide scripts for fine-tuning under script/ (❗Preferred). You may need to modify the DATA_DIR and OUTPUT_DIR paths in run_main.sh before running the scripts.
For example, you can fine-tune GOProteinGNN for contact prediction by running the following script:
sh ./script/run_contact.shYou can also use the running codes in run_downstream.py to write shell files with custom configurations.
run_downstream.py: support{ss3, ss8, contact, remote_homology, fluorescence, stability}tasks;run_stability.py: supportstabilitytask;
An example of fine-tuning GOProteinGNN for contact prediction (script/run_contact.sh) is as follows:
bash run_main.sh \
--model gcn-output/pretrained/GOProteinGCN/encoder \
--tokenizer "Rostlab/prot_bert" \
--output_file contact-GOProteinGCN \
--task_name contact \
--do_train True \
--epoch 5 \
--optimizer AdamW \
--per_device_batch_size 2 \
--gradient_accumulation_steps 8 \
--eval_step 50 \
--eval_batchsize 1 \
--warmup_ratio 0.08 \
--learning_rate 3e-5 \
--seed 3 \
--frozen_bert FalseArguments for the training and evaluation script are as follows,
--task_name: Specify downstream task. The script supports{ss3, ss8, contact, remote_homology, fluorescence, stability}tasks;--model: The name or path of a pre-trained protein language model checkpoint.--tokenizer: The name or path of a tokenizer.--output_file: The path to save fine-tuned checkpoints and logs.--do_train: Specify if you want to fine-tune the pretrained model on downstream tasks. Set this toFalseif you want to evaluate a fine-tuned checkpoint on the test set.--epoch: Number of epochs for training.--optimizer: The optimizer to use, e.g.,AdamW.--per_device_batch_size: Batch size per GPU.--gradient_accumulation_steps: The number of gradient accumulation steps.--eval_step: Number of steps to run evaluation on validation set.--eval_batchsize: Evaluation batch size.--warmup_ratio: Ratio of total training steps used for a linear warmup from 0 tolearning_rate.--learning_rate: Learning rate for fine-tuning--seed: Set seed for reproducibility--frozen_bert: Specify if you want to freeze the encoder in the pretrained model.
More detailed parameters can be found in run_main.sh. Note that the best checkpoint is saved in OUTPUT_DIR/.
Semantic similarity inference and binding affinity estimation are tasks from PROBE.
The code for PROBE can be found in src/benchmark/PROBE.
To validate GOProteinGNN on these two tasks, you need to:
- Configure paths in
src/benchmark/PROBE/extract_embeddings.pyto your pre-trained model and PROBE data accordingly. - Extract embeddings using pre-trained GOProteinGNN by running
src/benchmark/PROBE/extract_embeddings.py. - Change paths listed in
src/benchmark/PROBE/bin/probe_config.yamlaccordingly. - Run
src/benchmark/PROBE/bin/PROBE.py.
Detailed instructions and explanations of outputs can be found in PROBE.
The code for PPI can be found in src/benchmark/GNN_PPI, which was modified based on GNN-PPI.
To validate GOProteinGNN for PPI prediction:
- Configure paths in
src/benchmark/GNN_PPI/extract_protein_embeddings.pyto your pre-trained model and PPI data accordingly. - Extract embeddings using pre-trained GOProteinGNN by running
src/benchmark/GNN_PPI/extract_protein_embeddings.py. - Change paths listed in
src/benchmark/GNN_PPI/run.pyaccordingly. - Run
src/benchmark/GNN_PPI/run.py(training stage). - Run
src/benchmark/GNN_PPI/run_test.py(test stage).