MyoPy is an open-source Python package for the preprocessing and analysis of electrophysiological muscle signals. In its current state, it only allows for preprocessing raw data into epochs, estimating the EMG onset and extracting features from the EMG data for classification / machine learning.
To get the latest version of MyoPy clone the repository
https://github.com/k0ssmann/myopy
and install it via
python setup.py install
- Python>=3.11
- numpy>=1.26
- pandas>=2.2.0
- PyQt5>=5.15
- pyqtgraph>=0.13
MyoPy supports in its current state only input from tabular files. To show the basic functionalities of MyoPy, we'll use the example data provided in `datasets'. Let's start by importing the required modules for reading your EMG data from tabular files
from myopy.info import create_info
from myopy.io.tables import tables
from myopy.epochs import Epochs
from myopy.events import find_eventsIn the next step, we want to create an Info object that stores information such as the names of the EMG channels and the sample rate
ch_names = ['EMG1', 'EMG2', 'EMG3', 'EMG4']
sfreq = 20
info = create_info(ch_names, sfreq)As most EMG data stored in a tabular format isn't standardized, we have to provide information where in the file, i.e in which columns, our data is and, if available, in which columns our events are encoded (usually in TTL). Then we can create a Raw object from our data
For the provided dataset this would look like this
fname = './datasets/01-01.txt'
col_data = [19, 20, 21, 22]
col_events = [23, 24, 25, 26]
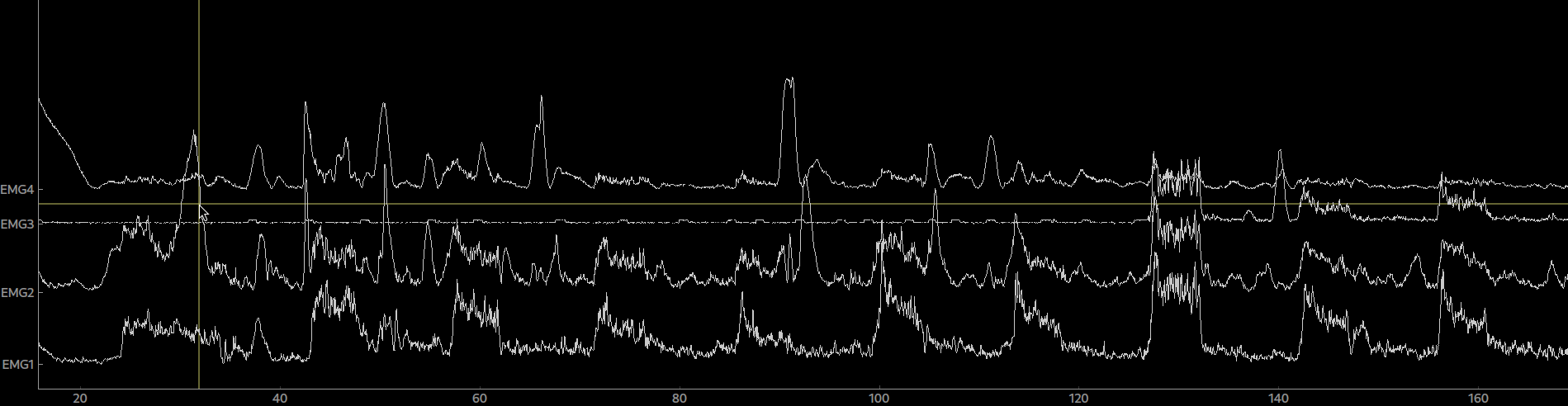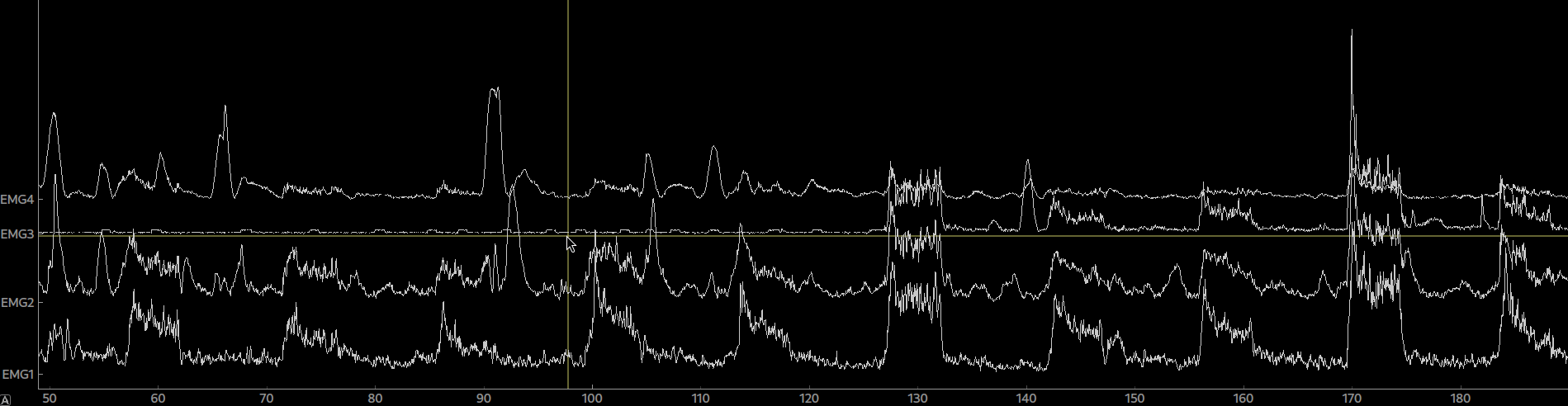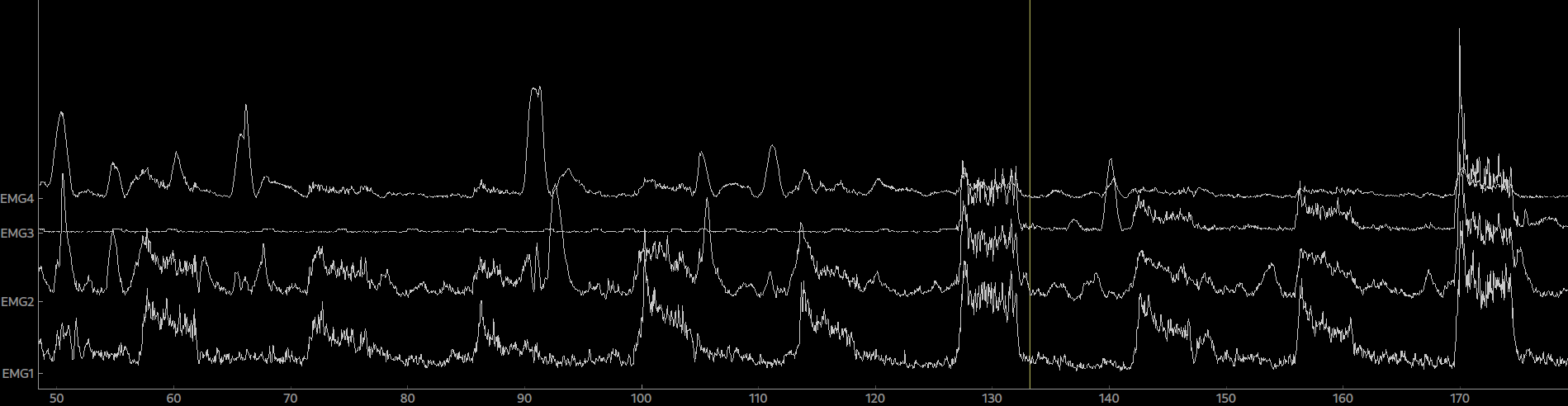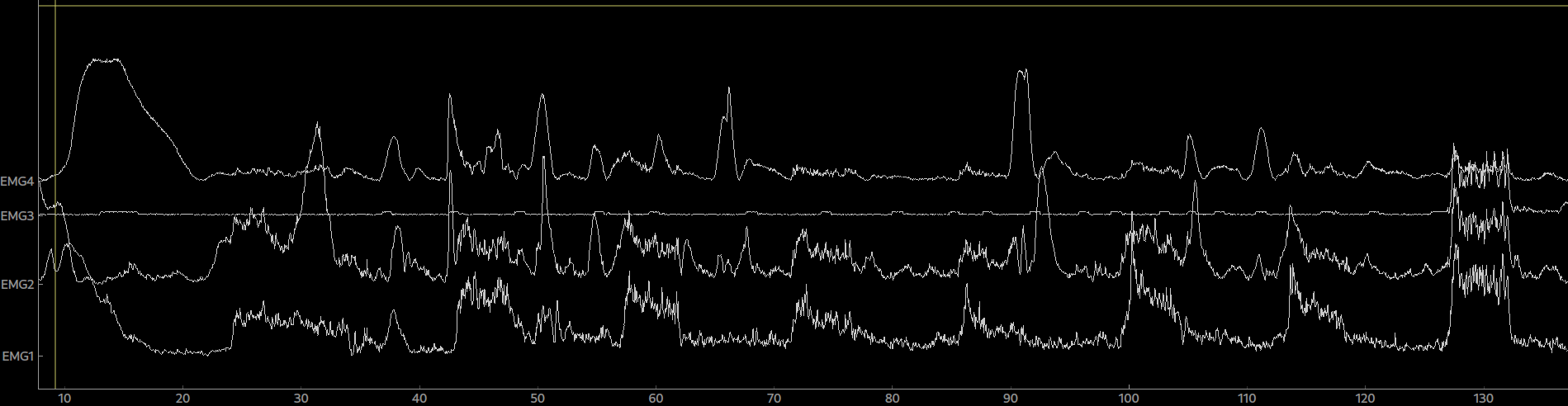
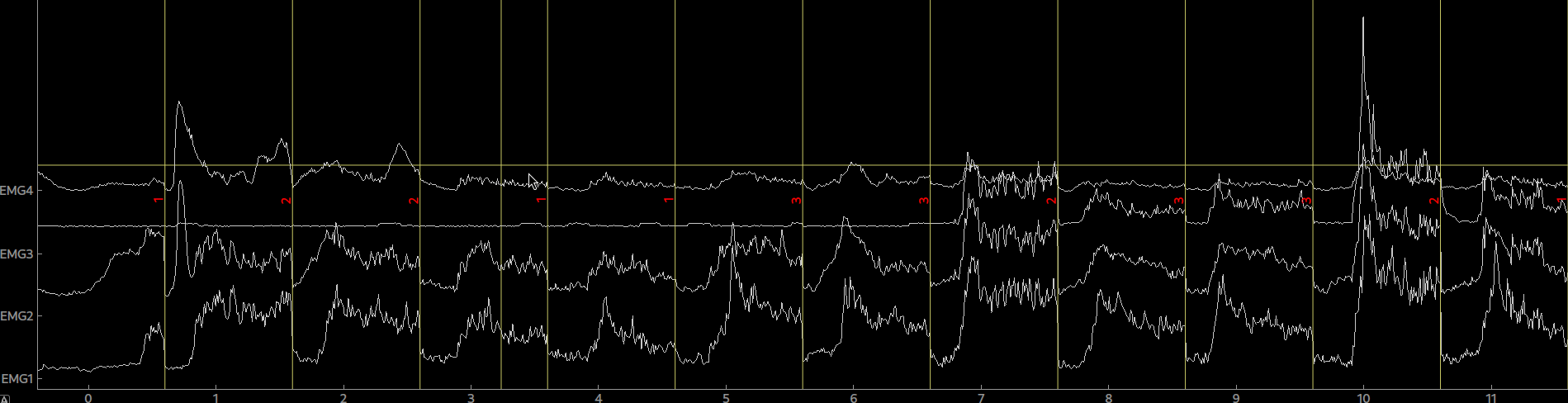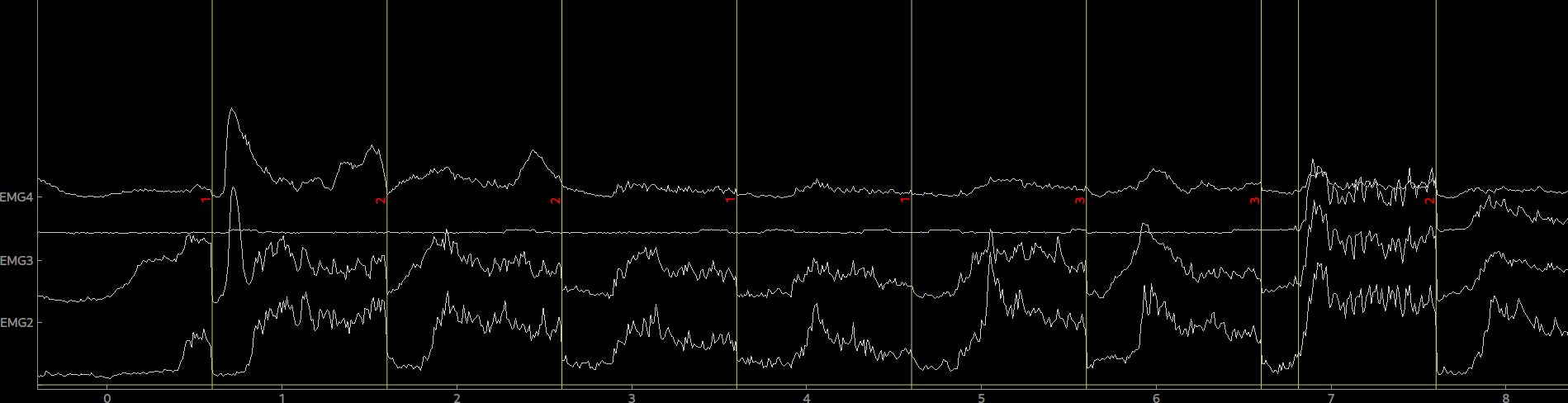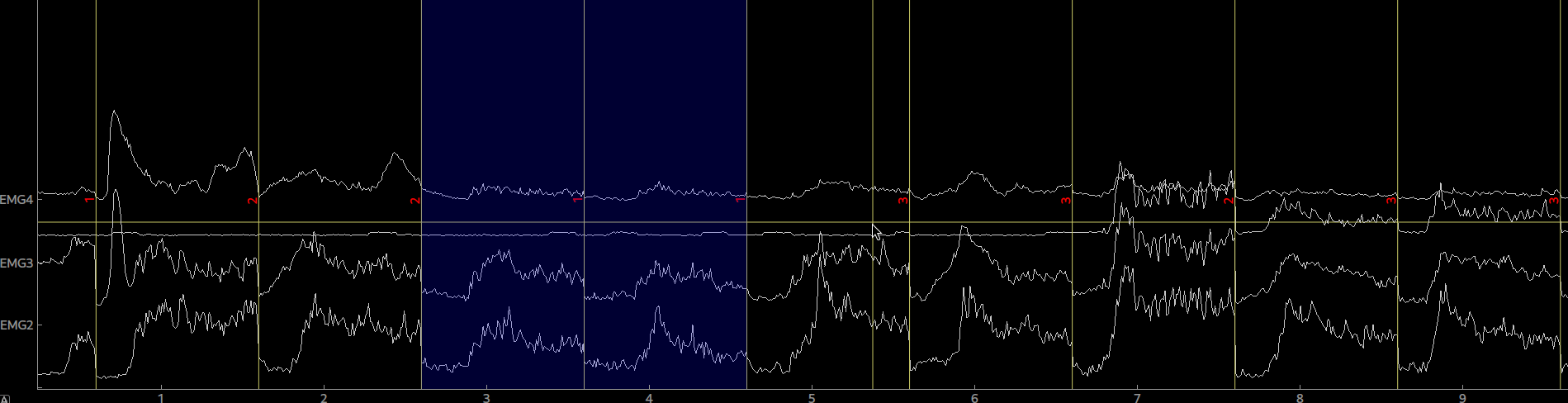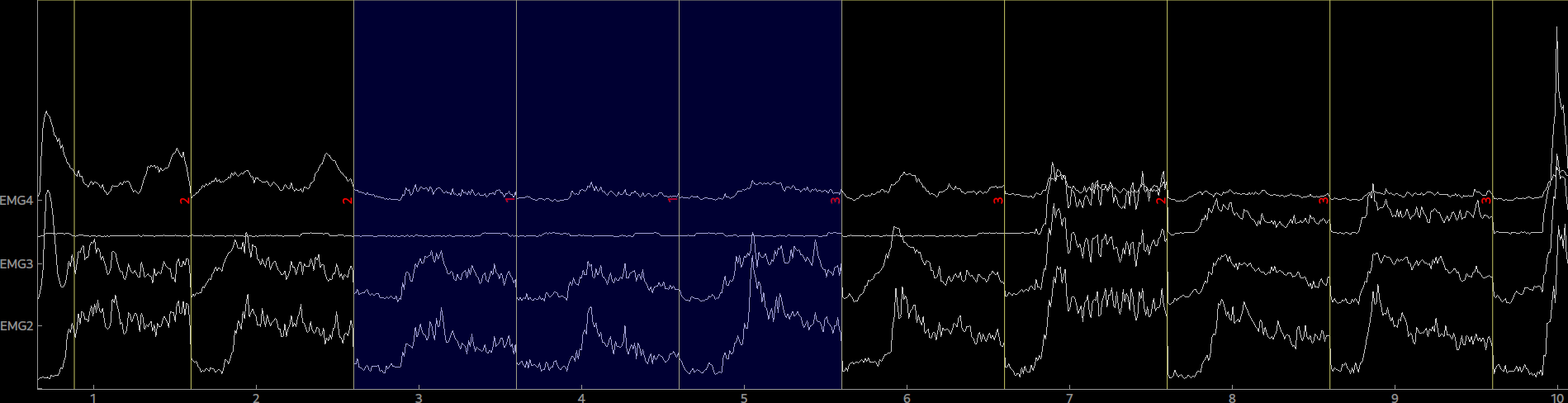
raw = tables.read_table(fname=fname, info=info, col_data=[19,20,21,22], col_events=[23,24,25,26], delimiter='\t')By calling plot(picks=[0,1,2,3]) on the raw object we can now plot our data.
To cut our data into segments (epoching) we have to create an Epoch object by first finding event onsets and specifying which events we want to see. In the example dataset the event IDs range from 0 - 7, however, only 1 - 6 contain event-correlated muscle signals.
event_ids = [1, 2, 3, 4, 5, 6]
events = find_events(raw)
epo = Epochs(raw, events=events, event_id=event_ids, picks=[0,1,2,3], tmin=0, tmax=5.0)Again, the epoched data can be plotted by calling plot() on the Epoch object. By clicking on an epoch you can mark it as bad and by calling drop_bads() you can remove them.
MyoPy is licensed under the MIT license.