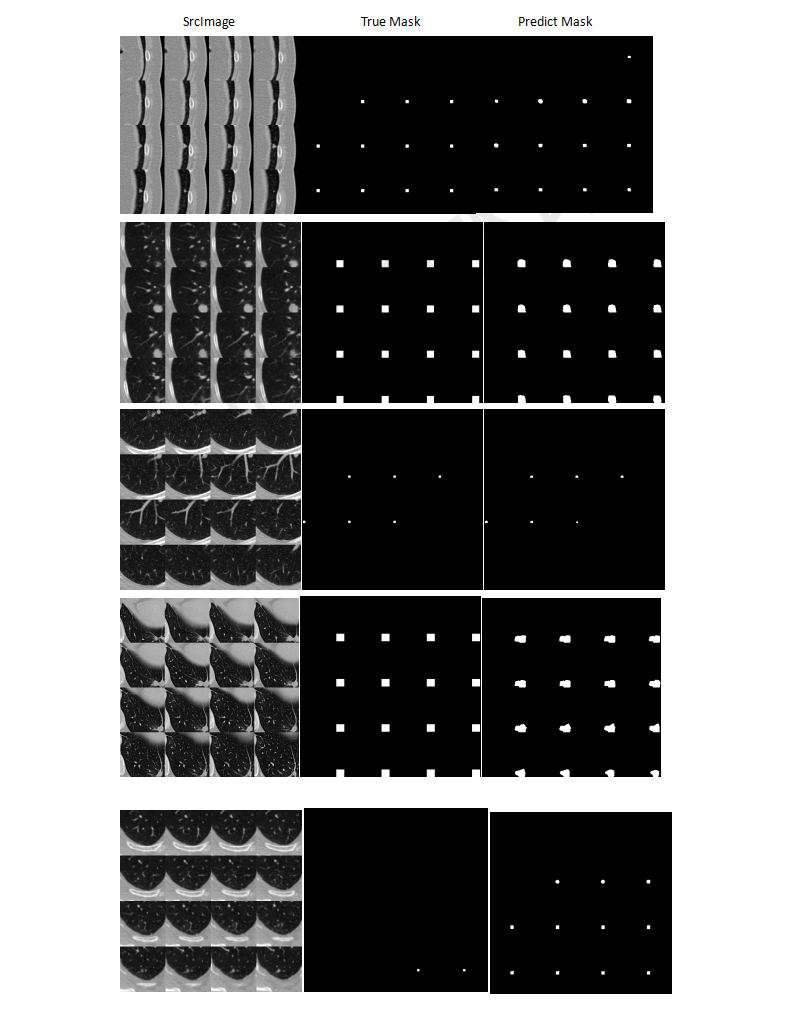
This is an example of the CT images lung nodule detection and false positive reduction from LUNA16-LUng-Nodule-Analysis-2016-Challenge
The following dependencies are needed:
- numpy >= 1.11.1
- SimpleITK >=1.0.1
- opencv-python >=3.3.0
- tensorflow-gpu ==1.8.0
- pandas >=0.20.1
- scikit-learn >= 0.17.1
1、Preprocess
nodule detection
- convert annotation.csv file to image mask file:run the LUNA_mask_extraction.py
- analyze the ct image,and get the slice thickness and window width and position:run the dataAnaly.py
- generate lung nodule ct image and mask:run the data2dprepare.py
- generate patch(96,96,16) lung nodule image and mask:run the data3dprepare.py
- save lung nodule data and mask into csv file run the utils.py,like this:G:\Data\segmentation\Image/0_161....
nodule classify
- convert candidates.csv file to nodule and not-nodule image(48,48,48):run the LUNA_node_extraction.py
- Augment the nodule image data: run the Augmain.py
- split data into train data(80%) and test data(20%):run the subset.py
- save lung nodule data and label into csv file like this:1,G:\Data\classify\1_aug/0_17.npy
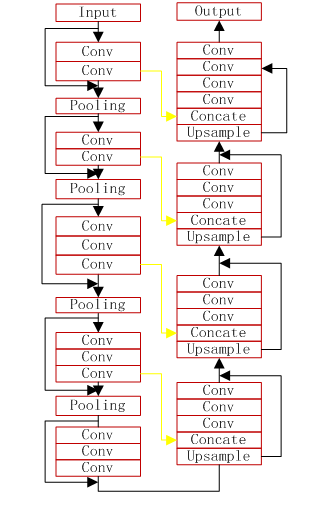
2、Nodule Detection
- the VNet model
- train and predict in the script of vnet3d_train.py and vnet3d_predict.py
3、False Positive Reducution
- the ResVGGNet model
- train and predict in the script of ResNet3d_train.py and ResNet3d_predict.py
4、download trained model
- i have shared the trained model of nodule detection and false positive reduction on here: https://pan.baidu.com/s/1I7zhzmPsTCbz0ZeIntNrUA ,password:orpm
1、Nodule Detection
- train loss and train accuracy
- the segment result
2、False Positive Reducution
- train loss and train accuracy
- ROC,Confusion Matrix and Metrics
- https://github.com/junqiangchen
- email: 1207173174@qq.com
- Contact: junqiangChen
- WeChat Number: 1207173174
- WeChat Public number: 最新医学影像技术