Congrats to everyone! We are glad to get 16th in this competition. We want to say thank to our team members and all participants in this competitions! Especially thank host to celebrate such an amazing competition! It's my first time to see hosts are so nice to help people facing issues in discussion, also post some great kernels to help us dive into this competition! Great job everyone!
5 cell-wise model, 3 image-wise model, 1 yolov5 model. All image-wise model receive green channels only.
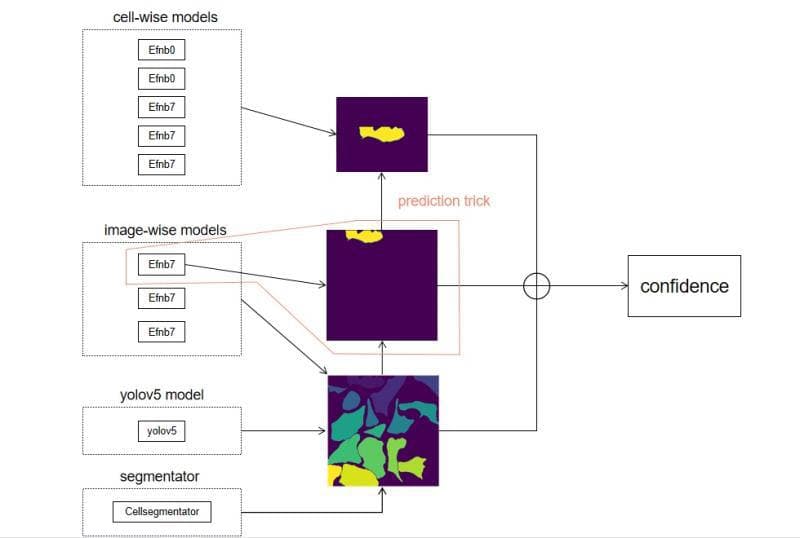
0.5 cell-wise model + 0.3 image-wise model + 0.1 yolov5 model + 0.1 image-wise model(predict the cell)
1.Efficientnet b0, external data included, pseudo labeling, epoch 10;
2.Efficientnet b0, external data included, pseudo labeling, epoch 1;
3.Efficientnet b7, external data included, epoch 1;
4.Efficientnet b7, epoch 1;
5.Efficientnet b7, external data included, green channel only, epoch 1.
1.Efficientnet b7, external data included, green channel only, epoch 20;
2.Efficientnet b7, green channel only, epoch 20;
My public kernel
3.Efficientnet b7, green channel only, input size 720, epoch 20;
@aristotelisch ‘s public kernel
Add padding to keep the cell tile’s width equals height, then resize to the input size, which can keep the cell tile’s width-height ratio unchanged, highly boosting our score.
Considering the cell-wise tile has unreliable class, we think pseudo labeling on cell may help. Here is our strategy:
1)Split datasets to two part;
2)Use one predict the other;
3)For the possibility of each class:
i)If confidence value>0.8, put this class into class list;
ii)If confidence value<0.2, if the answer is YES, we will put class 18 into class list;
v)Drop cell tiles without any class.
I have posted my public kernel about the ensemble of cell-wise and image-wise models. I’m glad to see that it helps. Nevertheless, in some cases, adding image-wise models may not improve the performance. Basically, if it does not work, you can try to tune down the weights of image-wise models. I joined VinBigData competition before, in which i use the weight of 0.95detection + 0.05image-wise model. Even the score went down when i tuned the weight of detection to 0.9. In most cases, more powerful the cell-wise model is, less improvement you can get by ensemble with image-wise models. It’s normal because the mAP focus on the cell-wise confidence anyway. Basically, if one want to do ensemble of cell-wise and image-wise models. Maybe you want to use multiplication first. Nevertheless, I think weighted average may help and easy to conduct. This idea is from my combine of detection and CNN. I think detection is to get bbox, but CNN may predict more accurate confidence value than detection. However, False positives becomes a main issue here. We will expect that image-wise model can help the performance a little bit and that is enough. In this competition, however, we do not have reliable cell-wise labels, so image-wise models may help a lot. If a person can solve the weak-label issue, they will get less improvement from image-level models, but the upper limit of score is relatively high
Here comes the interesting part--
Considering we don’t have the GT of the mask( they can only be obtained by CellSegmentator ), we think it is not easy to make our segmentation models more accurate than CellSegmentator . Nevertheless, yolov5 can not only detect the image into cell bbox but also give each bbox a confidence value. Considering all of these, we select yolov5 to our model zoo. However, considering the mAP is calculated by the sorting of confidence value, different location of cells in images may influence the confidence value for yolov5. So it is necessary to train other models to predict cell-tile data to do the ensemble.
Training: Green channel only, no external data, 20 epochs for basic training, 20 epochs for fine tuning.
Prediction:
1)Get the mid points of each cell by Cellsegmentator;
2)Do the KNN(N=1) with mid points predicted by yolov5, making cells and confidence values compared. When a cell mask has two bbox confidence values, choose the larger one. (like NMS)
3)Assign each cell with the confidence value given by yolov5. If KNN does not find bbox of yolov5 to compare, set the confidence value to 0. Finally, weighted average with our cell-level model. (Like WBF)
In this competition, the labels of cell tiles are not quite stable. Meanwhile, the image-wise model seems more robust because the training data of image-wise label is reliable. So, we mainly want to find a way to use image-wise model to predict the cell.
So we came up with a way, it goes like this:
1)Label a whole image using Host’s Segmentator, so we will get different cells in the image(1,2,3 …);
2)Choose a cell in interest, set all area except that to 0;
3)Resize the image to the size that image-wise model need;
4)Predict. Image-wise models(My public kernel )
I think it is a 'magic' part of our solution.
Thanks all from participants to host. It’s a such amazing journey for us. Hope to see you guys in HPAv3 :)
Our main training and inference codes are in Kaggle, so we kindly encourage you run the kernels in Kaggle to reproduce the result. Nevertheless, you can still get the code from this github repository if you want.
Concretely,
-
Run the data-preparing code to get the newest tfrec link for TPU training and generate the train.txt for yolov5.
https://www.kaggle.com/louieshao/get-gcs-path
https://www.kaggle.com/h053473666/hpa-yolo-txt -
Run the training code first to get the models prepared.
Yolov5:
https://www.kaggle.com/h053473666/hpa-yolov5-train-all
https://www.kaggle.com/h053473666/hpa-yolov5-train-all-fine
Image-wise models:
https://www.kaggle.com/louieshao/hpa-classification-efnb7-train-tfrec
Cell-wise models:
https://www.kaggle.com/louieshao/hpa-training-tpu
- Run the inference kernel
https://www.kaggle.com/h053473666/hpa-yolov5-ensemble-final