This repository serves to store code and an executable for Windows to process the output from maxquant for complexome profiling experiments. The maxquant file is processed in 5 steps. In the first step unwanted columns are removed and proteins not of interest (currently these proteins contain the words "REV" or "CON" in the 'Majority protein IDs' column) are filtered away. In the second step proteins are queried in batches to uniprot and per protein the gene name, protein name, organism name, the cell compartment, a uniprot specific protein hyperlink and a hyperlink to the string database are retrieved, if they are available. The user can specify which elements should be retrieved from uniprot. In the third step each protein is compared with the proteins from the mitocarta 'database' and in a column it is denoted if the protein is present in the human or mouse mitocarta database. In order to know which proteins are present in mitocarta the gene name from uniprot is used. In the fourth step the different samples are clustered based on hierarchical clustering. Additionally, (globally) all samples together are clustered. In the fifth and final step the processed output is written away to an excel file with the 2 different sheets where the first sheet contains the proteins with information as well as the accompanying complexome profiling data(conditional formatting has been applied on these columns) and the second sheet contains the filtered away proteins.
This program uses a settings file in which the user can disable individual steps, change output behaviour and more. Whenever the user enters invalid parameters, depending on the parameter in the worst case the program will halt and show an error message. Lastly, whenever the uniprot step is disabled the mitocarta step will also not be executed, because the gene name per protein from uniprot is used as input for the mitocarta step.
On the highest level the user is able to disable or enable each individual step. For the user it doesn't make much sense to disable the first or fifth step, but for the programmer who made this it is really convenient to be able to disable each indivual step. For the first step the user is able to alter which columns and rows/proteins should be retained from the maxquant file. Proteins who do not meet the criterion will be written to a separate excel sheet. For the second step the user is able to choose globally which elements should be queried from uniprot. In order to query uniprot several url's are needed, whenever these url's result in bad requests the usr should replace these with an updated url. Additionally, the user is able to change how many proteins are queried per batch and how much idle time there should be between batches. For the third step the user is able to correct the location of the mitocarta database whenever this changes to a newer version, change which columns should be evaluated and enable or disable which columns the program evaluates. For the fourth step the user is able to change the metric(see [this](https://docs.scipy.org/doc/scipy/reference/generated/scipy.spatial.distance.pdist.html#scipy.spatial.distance.pdist) page for an explanation of different metrics) or method(see [this](https://docs.scipy.org/doc/scipy/reference/generated/scipy.cluster.hierarchy.linkage.html) page for an explanation of different methods) that is used with hierarchical clustering. For the fifth and final step the user is able to change the name or location(differently than the current directory/folder) of the excel file. Additionally, the user is able to change the order of the columns which were originally in the maxquant file except for the sample columns which are sorted alphabetically.
In order to run the executable you first need to download the executable and user defined settings file. First step, go to the releases web page. Second step, download the executable file from the newest release and the 'maxquant_settings.json' file.
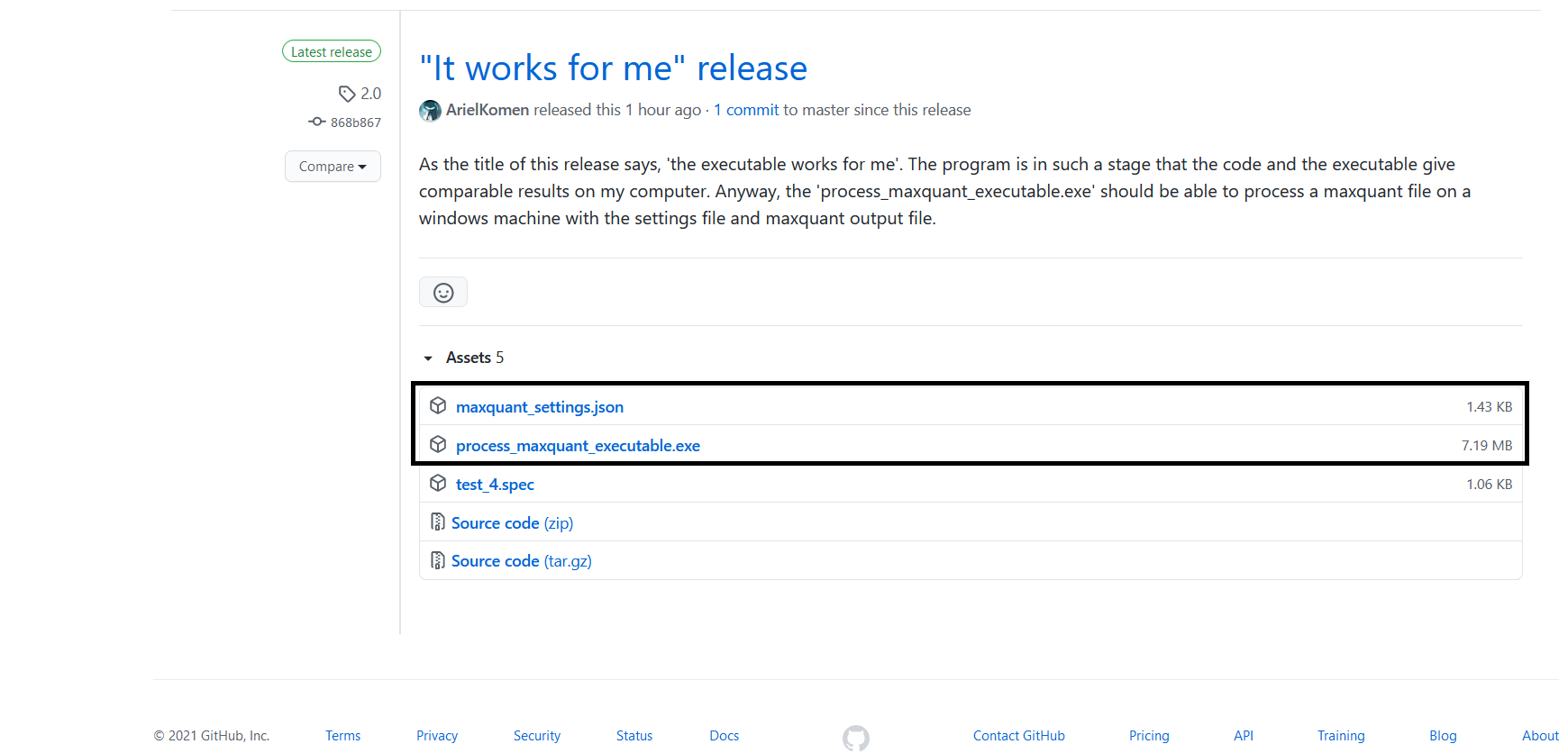
Once you have downloaded the executable and settings file place them together in a folder.
This folder should contain (1) the maxquant file you wish to process, (2) the settings file and (3) the executable.
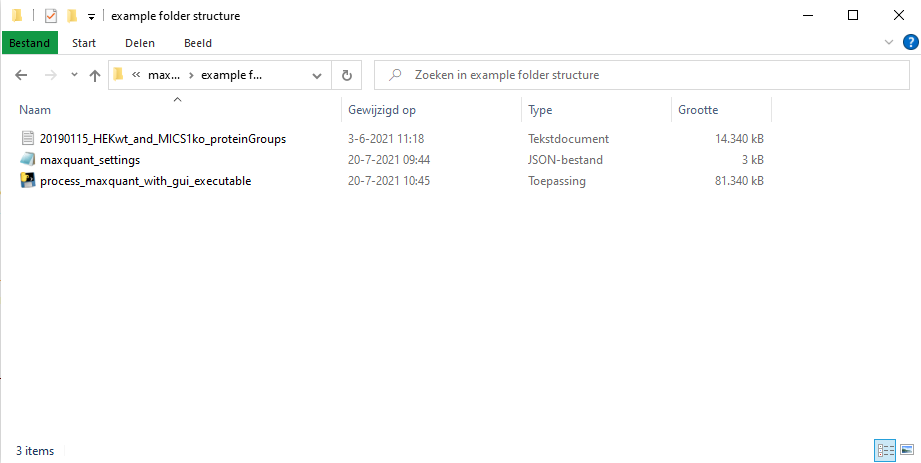
Now is a good moment to open the settings file and based on the description of the user defined parameters down below change parameters in order to change the behaviour of how the maxquant output is processed.
An example setup:
Now the program is ready to run. Start the program by executing the executable which will start a shell script and after 1 second a Graphical User Interface (GUI) will appear.
In order to run the program it needs the settings file and maxquant file. You can select the maxquant output file by clicking 'Select maxquant file' which will open a file selection menu
where you can select the file. The file path to the maxquant file is put in the textbox above the 'Select maxquant file' button.
The settings file is selected in a similar way as the maxquant file, but you should select the maxquant file by clicking 'Select the settings file' button.
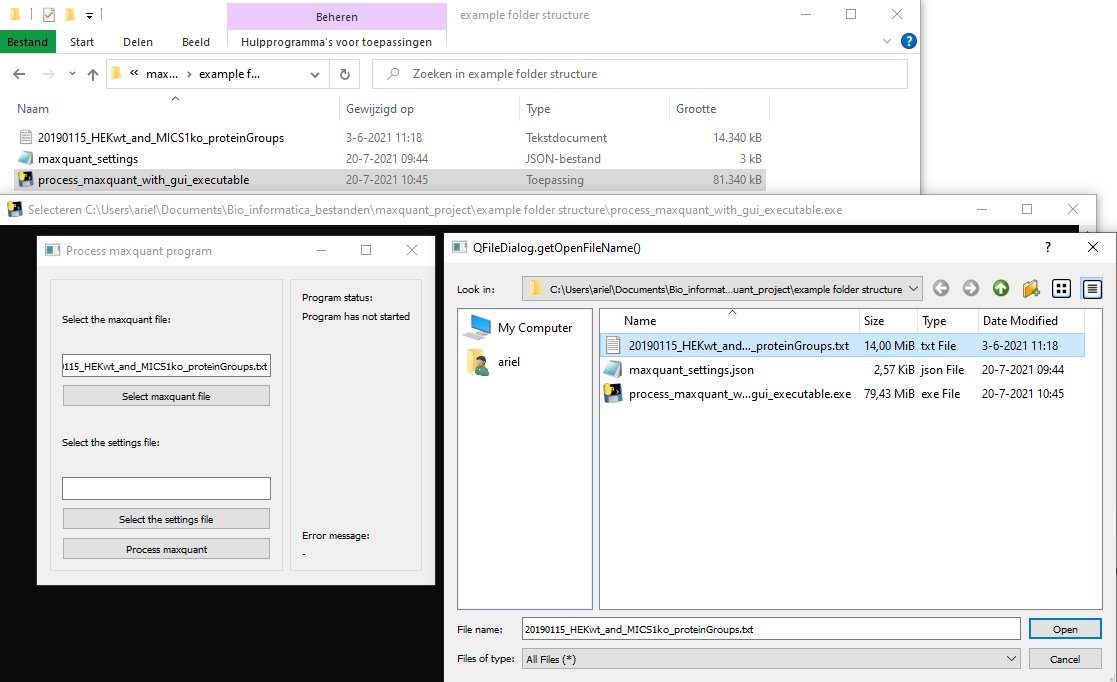
When both files have been selected the program can be execute by clicking 'Process maxquant'. The GUI will inform you through status messages and whenever something goes wrong an error message will appear.
1. steps_dict -> which steps would you like to execute? 1 means that the step is executed and 0 means the step is not executed. The program expects a 1 or 0 and nothing else will work. 1. filtering_step 2. uniprot_step 3. mitocarta_step 4. clustering_step 5. make_excel_file_step-
filtering_step -> parameters for the filtering step
- EXACT_MATCHES -> Elements in this list should be retained from the maxquant file.
- CONTAINS -> Columns in the maxquant file which contain elements from this list should be retained.
- PROTEIN_FILTERS -> Rows/proteins containing elements from this list will not be retained(will be written to a separate excel sheet)
-
uniprot_step -> parameters for querying uniprot
- uniprot_options -> a dictionary similiar to steps_dict which enables the user to define which elements should be retrieved from uniprot. These parameters shouldn't be anything else than 1 or 0
- get_gene_name
- get_protein_name
- get_organism_name
- get_uniprot_hyperlink
- get_cell_compartment
- get_string_linkout
- uniprot_base_url -> This url is the basis for querying uniprot
- uniprot_request_url -> This url has parameters necessary to query uniprot and is combined with 'uniprot_base_url'.
- request_idle_time -> In order to prevent being blacklisted between batches the program waits a few seconds. How many seconds should the program wait? Change this parameters at you own risk
- batch_amount -> Uniprot is queried in batches of size n where n shouldn't be larger than 100.
- uniprot_protein_base_url -> The hyperlink to each protein of the uniprot database has a generic part + protein name, this url encodes this generic path.
- known_gene_names -> In order to retrieve the gene name from uniprot several keys are known, this list contains the known gene names.
- known_protein_names -> similar to known_gene_names but for protein_names.
- string_linkout_parameters -> dictionary containing information to query the uniprot mapping service to get string linkouts
- string_base_url -> The base url to query string
- uniprot_mapping_service_url -> the base url to query the uniprot mapping service
- regex_pattern -> a regex pattern is needed to locate proteins with a suffix like "-2" which needs to be removed from proteins in order to query them to the mapping service of uniprot.
- uniprot_options -> a dictionary similiar to steps_dict which enables the user to define which elements should be retrieved from uniprot. These parameters shouldn't be anything else than 1 or 0
-
mitocarta_step -> parameters for querying the mitocarta database
- mitocarta_human_ftp_link -> A link to the human mitocarta excel file/database
- mitocarta_mouse_ftp_link -> A link to the mouse mitocarta excel file/database
- human_sheet_name -> the sheet name that should be used for the human excel file
- mouse_sheet_name -> the sheet name that should be used for the mouse excel file
- mitocarta_symbol_column -> What is the main column in the mitocarta database to examine whether the protein is in the mitocarta database?
- mitocarta_additional_symbol_column -> Proteins have synonyms, which column in the mitocarta database contains synonyms for the gene names?
- evaluate_symbol_column -> Should the symbol column be evaluated? Expecting 0 or 1.
- evaluate_additional_symbol_column -> Should the additional symbol column be evaluated? Expecting 0 or 1.
-
clustering_step -> parameters for clustering the complexome profiling samples
- method -> method for how the clustering is performed. Possible options are: 'single', 'complete', 'average', 'weighted', 'centroid', 'median' or 'ward'.
- metric -> which distance metric should be used. Possible distance metrics are: 'braycurtis', 'canberra', 'chebyshev', 'cityblock', 'correlation', 'cosine', 'dice', 'euclidean', 'hamming', 'jaccard', 'jensenshannon', 'kulsinski', 'mahalanobis', 'matching', 'minkowski', 'rogerstanimoto', 'russellrao', 'seuclidean', 'sokalmichener', 'sokalsneath', 'sqeuclidean', 'yule'.
-
make_excel_file_step -> parameters for writing away the data into an excel file
- excel_file_name -> The name of the final excel file. Can also be a absolute path to a desired location.
- output_column_order -> What should the order be of the first columns of the excel file? Column names not found in the maxquant file will not be present in the excel file.