How to combine feature-based function and Graph based Function?
mrbeann opened this issue · comments
I'm solving a problem that can be formulated as
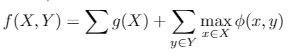
The second part is a normal facility location problem, while the first one is a function associate with each data in X (e.g. the value of the first feature).
And I try to use the MixtureSelection with it as follows,
def feature_based_function(X):
return X[:, -1].sum().sum()
def facility_location_function(X):
return X[:, :-1].max(axis=0).sum()
v = np.random.uniform(0.5, 1.0, (50, 1))
similarity = np.random.uniform(0.5, 0.9, size=(50, 50))
n = 10
x = np.concatenate([v, similarity], axis=1)
weight = [1, 1]
func1 = CustomSelection(n, feature_based_function)
func2 = CustomGraphSelection(n, facility_location_function, metric='precomputed')
func3 = MixtureSelection(n, [func1, func2], weights=weight, optimizer='naive').fit(x)
selected = func3.transform(x)But it seems this can not generate a good subset, is there any problem?
Howdy
What do you mean when you say that "it can not generate a good subset"? I can't remember apricot correctly handles a mixture of a graph- and a feature-based function, because a feature-based custom function requires access to the feature values and the graph-based one requires access to rows of an adjacency matrix. However, your feature based function doesn't actually seem like a submodular function. Can you clarify what it is you're doing there? If it's just adding a single value, then it's not submodular.
Thanks for your reply.
- As for "it can not generate a good subset" I use several variants (random select, or only consider feature or similarity part) and test it with the objective function.
- The feature-based function is to select the max sum from a set. It satisfies
$f(A U x) - f(A)=f(B U x)-f(B)$ . For physical meaning, it means each item has a value, I need to maximize the value of selected set. - I understand these two kinds of function needs different input, so I slice the input for a different function, is it okay?
- It would be helpful if you were more specific. You test it with the objective function and see what? What were you expecting to see?
- Do you mean that you have a scalar value that is, essentially, a bonus for choosing an item when optimizing the facility location function? Put another way: without the facility location component, you would just choose the k items with the highest value?
- A function passed into
CustomGraphSelectionshould take in rows of an adjacency matrix, whereas a function passed intoCustomSelectionshould take in rows of a feature matrix (the normal format for ML problems). This is because feature-based functions calculate their gain using the feature values directly, whereas graph-based functions calculate the gain using a similar matrix (which can be calculated from feature values). You might have better luck trying to implement a custom selection object instead of passing a custom function into a selection wrapper (https://github.com/jmschrei/apricot/blob/master/tutorials/4.%20Defining%20Custom%20Functions.ipynb under "Writing your own selection objects"). That way you can store both the pre-computed adjacency matrix AND the feature matrix.
If what you want is the first feature (column) to be used in a feature-based manner and the other columns to be used to calculate an adjacency matrix, I'd start off with something like this:
from apricot import BaseSelection
class SkeletonSelection(BaseSelection):
# If you're defining a feature-based function you should inherit from BaseSelection
# If you're defining a graph-based function and want to make use of the built-in
# similarity calculations, you should inherit from BaseGraphSelection, which itself
# inherits from BaseSelection.
def __init__(self, n_samples, alpha, initial_subset=None, optimizer='two-stage',
optimizer_kwds={}, n_jobs=1, random_state=None,
verbose=False):
# If defining a graph-based function, you should also include a metric
# parameter, with the default in apricot being "metric='euclidean'".
# You will also need to pass the metric into the call to `super` below
# by adding "metric=metric" anywhere.
# This function must call `__init__` of the base class, as below.
# Other custom arguments, such as hyperparameters for the underlying
# submodular function, should be stored in this method.
self.alpha = alpha # This will control the weight between the two components
super(SkeletonSelection, self).__init__(n_samples=n_samples,
initial_subset=initial_subset, optimizer=optimizer,
optimizer_kwds=optimizer_kwds, n_jobs=n_jobs, random_state=random_state,
verbose=verbose)
def _initialize(self, X):
# This function includes any logic that should be executed before the
# selection process begins, such as initializing cached values and
# setting them to values from the initial subset.
X_pairwise = numpy.corrcoef(X[:, 1:].T) # or whatever your similarity function is
self.current_values = numpy.zeros(X.shape[1] - 1)
super(SkeletonSelection, self)._initialize(X)
def _calculate_gains(self, X, idxs=None):
# This function returns the gain in the objective function that each
# item in X[idxs] would return. The returned vector should be of
# length len(idxs) or len(self.idxs) if idxs is None. The value of
# gains[i] should be the gain associated with adding X[idxs[i]] to
# the selected set.
idxs = idxs if idxs is not None else self.idxs
gains = X[idxs][:,0] + self.alpha * numpy.maximum(X_pairwise[idxs], self.current_values[idxs]).sum()
return gains
def _select_next(self, X, gain, idx):
# This function takes in a single example, X, the corresponding gain
# of adding this element to the selected set, and the index of this
# example in the full data set, and updates the cached values accordingly.
self.current_values = numpy.maximum(self.current_values, X_pairwise[idx])
super(SkeletonSelection, self)._select_next(X, gain, idx)I haven't debugged this, but hopefully you can figure out how to get it to work for your application. Let me know if you have any more questions.
- the final object is
def obj(X):
weight[1] * facility_location_function(X) + weight[0]*feature_based_function(X)And I try two variants
# just the feature
func1 = CustomSelection(10, feature_based_function, optimizer='naive')
func1.fit(x)
selected = func1.transform(x)
print('first', obj(selected))
# just the distance
func2 = CustomGraphSelection(10, facility_location_function, metric='precomputed', optimizer='naive')
func2.fit(x[:, :-1])
selected = func2.transform(x)
print('second', obj(selected))After testing it 100 times, I get the mean objective as,
both: 180.82156881211816
first: 180.8519182052127
second: 180.80307370127528
I also try different weights, but the mixture method never performs best.
2. Yes.
3. I read it, and try to use the simple (but slow) method to implement it. I'll try this kind of implementation.
I have tried this method, but it seems not to work properly.
class SkeletonSelection(BaseSelection):
def __init__(self, n_samples, alpha, initial_subset=None, optimizer='two-stage',
optimizer_kwds={}, n_jobs=1, random_state=None,
verbose=False):
self.alpha = alpha # This will control the weight between the two components
super(SkeletonSelection, self).__init__(n_samples=n_samples,
initial_subset=initial_subset, optimizer=optimizer,
optimizer_kwds=optimizer_kwds, n_jobs=n_jobs, random_state=random_state,
verbose=verbose)
def _initialize(self, X):
self.X_pairwise = X[:, :-1] # or whatever your similarity function is
self.current_values = np.zeros(X.shape[1]-1)
super(SkeletonSelection, self)._initialize(X)
def _calculate_gains(self, X, idxs=None):
idxs = idxs if idxs is not None else self.idxs
gains = X[idxs][:, -1] * self.alpha[0] + \
self.alpha[1] * np.maximum(self.X_pairwise[idxs], self.current_values[idxs]).sum()
return gains
def _select_next(self, X, gain, idx):
print(self.current_values.shape)
self.current_values = np.maximum(self.current_values, self.X_pairwise[idx])
super(SkeletonSelection, self)._select_next(X, gain, idx)
fun1 = SkeletonSelection(n, alpha=weight)
selected = fun1.fit_transform(X)Here I use the same data generated before. That X is a (n*(n+1)) matrix, the first (n*n) is the pre-computed distance and the last column is the bonus. But this will raise an error
self.current_values = np.maximum(self.current_values, self.X_pairwise[idx])
ValueError: operands could not be broadcast together with shapes (51,) (50,)
I have checked the code, is due to self.current_values in the BaseSelection is set with the dimension of X. Is there any idea how to deal with it?
It looks like the error is coming from super(SkeletonSelection, self)._initialize(X) rewriting the self.current_values variable. If you move self.current_values = np.zeros(X.shape[1]-1) that issue should be fixed.
what do you mean by "move self.current_values = np.zeros(X.shape[1]-1)"?
Move that line down one to be under the one starting super(...
Move that line down one to be under the one starting
super(...
I have tried this before, but it didn't work and raise a similar error.
self.alpha[1] * np.maximum(self.X_pairwise[idxs], self.current_values[idxs]).sum()
ValueError: operands could not be broadcast together with shapes (49,50) (49,)
Like I mentioned, I did not debug the code, and don't have time to do so right now. I'd encourage you to spend time looking at the apricot code yourself and seeing if you can figure it out. You'll only need to look at BaseSelection and the code I just wrote, so that should make it easier.
Thanks, I'll try to reimplement it with the BaseSelection.
A final question is am I do the right thing? I mean given the objective function and the feature-based function (as mentioned earlier in point 1 and 2). I should be able to see where apricot can find better results?
Yes, I think you're doing the right thing. You just need to make sure you evaluate the quality of the subset using the same function, regardless of how you chose the subset. Another thought I had is that, if you're just adding in a "bonus" for each item selected, you might find it easier to modify the FacilityLocationSelection object to take in a vector of "bonuses", one for each example, and add that value to the gain for each item. I don't think you need an actual feature-based function for what you're trying to do.
If you copy/paste the facility location function implementation I have, I think that all you'd need to do is change https://github.com/jmschrei/apricot/blob/master/apricot/functions/facilityLocation.py#L206 to also take in a vector of "bonuses" and save that using self.bonuses = bonuses, and then I think you would only have to modify https://github.com/jmschrei/apricot/blob/master/apricot/functions/facilityLocation.py#L309 to be return gains + self.bonuses[idxs]. When using this new object, you'd pass in the precomputed adjacency matrix (without the bonus feature in it at all) to the fit function as normal, but you'd pass the bonus feature in as a single vector when creating the object, e.g.
bonus = np.random.uniform(0.5, 1.0, (50, 1))
similarity = numpy.corrcoef(numpy.random.normal(0, 1, size=(50, 10)).T)
model = NewFacilityLocationSelection(10, metric='precomputed', bonus=bonus)
model.fit(similarity)Again, I haven't debugged that, but it might be faster for you to do that than try to reimplement something using the SkeletonSelection.
Thank you for your kindly reply, this solution sounds much better than mine.