This repo is inference for image-based orphan protein analysis in the paper Global organelle profiling reveals subcellular localization and remodeling at proteome scale. This analysis is by James Burgess and Chad Liu.
It copies the cytoself repo for training a Cytoself representation learning model. We modify the training a little bit (see next section), and then write some inference code for comparing representations of orphan proteins to representations from Opencell. In this Readme, the content after the section called cytoself is from the README of the original project.
- changed
requirements.txtfor torch and torchvision. Previously it was (torch>=1.11) but I was getting some error in the torch Upsample layer. I think it was some issue with torch 2.0, so I set it totorch==1.13.1andtorchvision==0.12. - scripts to train cytoself from scratch in
scripts - added the 10
.npyfile and 10.csvfiles fromhttps://github.com/royerlab/cytoself/tree/mainand put them indata/opencell_crops(which is obviously not comitted to the repo). - In the run scripts, I save the
train_argsandmodel_args. This is so that I can more easily reconstruct the model in the inference scripts. - in the
cytoself/datamanager/opencell.pyI add a step to the end of the dataloading of the dataloader. This saves the test dataset - it's indices and its crops into separate files. This is useful for doing analysis afterwards. You can load the embeddings and these labeles or images. It's necessary for doing inference with a new dataset. That puts data files in 'data/test_dataset_metadata/` - TODO: include the
data/cz_infectedcell_finalwellmapping.csvin the eventual repo.
The new inference code for is in inference/.
python inference/load_inf_data.pysaves stacks as max-intensity projs, and does some fov-level normalizations. Saved toresults/load_inf_data.python inference/nuclear_segmentation.py. Saves masks toinference/nuclear_segmentation/all_masks.pt. Issue is that if you take only a subset of these images, then you overwrite the pt file with only these images (this is not true of other steps)python inference/crop.py. For each segmented nucleus in the fov, take a crop around it. There is aVERSIONparameter that controls how normalization is done. IfVERSION==0(recommended), then do[0,1]normalization within each crop, treating nucleus and target channel independently. IfVERSION==1then normalize the FOV before cropping. IMO this is worse because, for example, you can get very bright (e.g. due to mitosis I guess), so doing normalization at the whole-FOV level makes the rest of the image very dim. If you normalize at the crop level, then only the abnormally bright areas are affected. Results saved ininference/results/crop/ascrops_v0forVERSION==0orcrops_v1forVERSION==1. Also the crop-metadata is saved tocrops_meta.csv, which has the fov filename, the centroid coords (in the fov-space) of the nucleus that is centered in this crop, and some other stuff.python inference/get_crop_features.pyloads the pretrained cytoself model. For model name, for example20240129_train_all, features saved toinference/results/get_crop_features/results/20240129_train_all/ckpt_None/. Need to define the pretrained models in the bottom of the script. Also an option to create features for rotated versions of the crops, which should give better robustness overall according to this paper. (you have to make sure theVERSIONmatches what was used in the cropping - sorry, this should have been handled automatically)python inference/compare_opencell_targets.pygets the 'consensus embeddings' for each protein by averaging over the crop representations for that protein. It does it for opencell and orphans, and makes a distance matrix for all proteins.
cytoself in pytorch implementation. The original cytoself implemented in tensorflow is archived in the branch cytoself-tensorflow.
Note: Branch names have been changed. cytoself-pytorch -> main, the previous main -> cytoself-tensorflow.
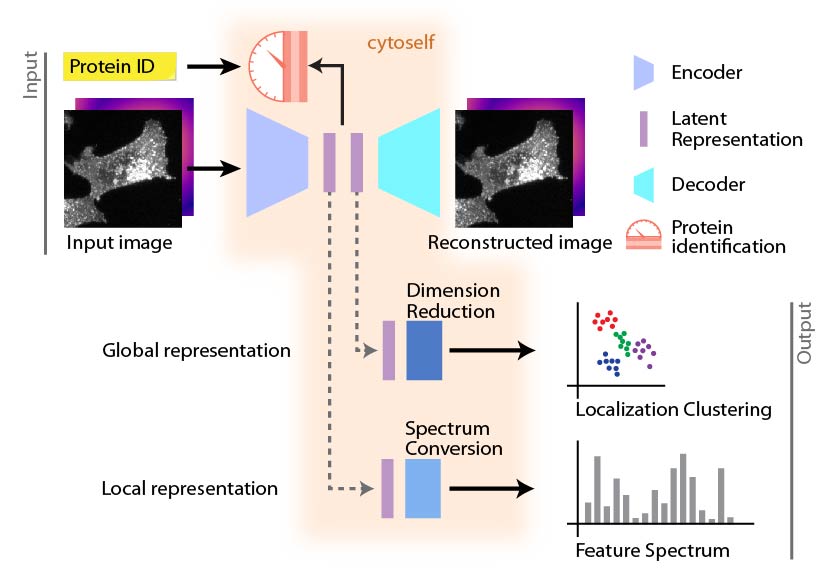
cytoself is a self-supervised platform for learning features of protein subcellular localization from microscopy images [1]. The representations derived from cytoself encapsulate highly specific features that can derive functional insights for proteins on the sole basis of their localization.
Applying cytoself to images of endogenously labeled proteins from the recently released OpenCell database creates a highly resolved protein localization atlas [2].
[1] Kobayashi, Hirofumi, et al. "Self-Supervised Deep-Learning Encodes High-Resolution Features of Protein
Subcellular Localization." Nature Methods (2022).
https://www.nature.com/articles/s41592-022-01541-z
[2] Cho, Nathan H., et al. "OpenCell: Endogenous tagging for the cartography of human cellular organization."
Science 375.6585 (2022): eabi6983.
https://www.science.org/doi/10.1126/science.abi6983
cytoself uses images (cell images where only single type of protein is fluorescently labeled) and its identity information (protein ID) as a label to learn the localization patterns of proteins.
Recommended: create a new environment and install cytoself on the environment from pypi
(Optional) To run cytoself on GPUs, it is recommended to install pytorch GPU version before installing cytoself following the official instruction.
conda create -y -n cytoself python=3.9
conda activate cytoself
# (Optional: Install pytorch GPU following the official instruction)
pip install cytoselfMake sure you are in the root directory of the repository.
pip install -e .Install development dependencies
pip install -r requirements/development.txtDownload one set of the image and label data from Data Availability.
is available.
from cytoself.datamanager.opencell import DataManagerOpenCell
data_ch = ['pro', 'nuc']
datapath = 'sample_data' # path to download sample data
DataManagerOpenCell.download_sample_data(datapath) # donwload data
datamanager = DataManagerOpenCell(datapath, data_ch, fov_col=None)
datamanager.const_dataloader(batch_size=32, label_name_position=1)A folder, sample_data, will be created and sample data will be downloaded to this folder.
The sample_data folder will be created in the "current working directory," which is where you are running the code.
Use os.getcwd() to check where the current working directory is.
9 sets of data with 4 files for each protein (in total 36 files) will be downloaded.
The file name is in the form of <protein_name>_<channel or label>.npy.
*_label.npyfile: Contains label information in 3 columns, i.e. Ensembl ID, protein name and localization.*_pro.npyfile: Image data of protein channel. Size 100x100. Images were cropped with nucleus being centered (see details in paper).*_nuc.npyfile: Image data of nucleus channel. Size 100x100. Images were cropped with nucleus being centered (see details in paper).*_nucdist.npyfile: Data of nucleus distance map. Size 100x100. Images were cropped with nucleus being centered (see details in paper).
from cytoself.trainer.cytoselflite_trainer import CytoselfFullTrainer
model_args = {
'input_shape': (2, 100, 100),
'emb_shapes': ((25, 25), (4, 4)),
'output_shape': (2, 100, 100),
'fc_output_idx': [2],
'vq_args': {'num_embeddings': 512, 'embedding_dim': 64},
'vq_args': {'num_embeddings': 512},
'num_class': len(datamanager.unique_labels),
'fc_input_type': 'vqvec',
}
train_args = {
'lr': 1e-3,
'max_epoch': 1,
'reducelr_patience': 3,
'reducelr_increment': 0.1,
'earlystop_patience': 6,
}
trainer = CytoselfFullTrainer(train_args, homepath='demo_output', model_args=model_args)
trainer.fit(datamanager, tensorboard_path='tb_logs')from cytoself.analysis.analysis_opencell import AnalysisOpenCell
analysis = AnalysisOpenCell(datamanager, trainer)
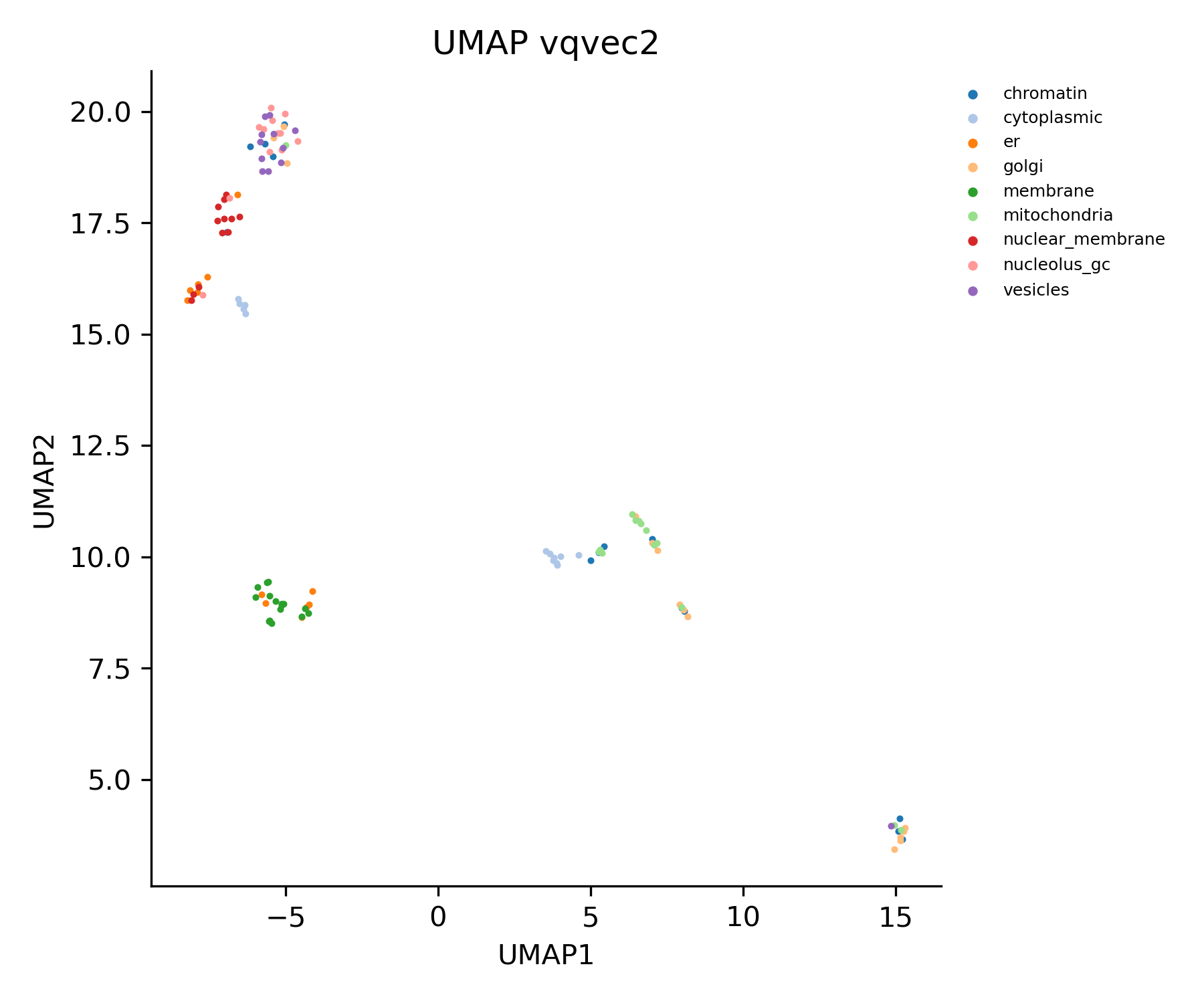
umap_data = analysis.plot_umap_of_embedding_vector(
data_loader=datamanager.test_loader,
group_col=2,
output_layer=f'{model_args["fc_input_type"]}2',
title=f'UMAP {model_args["fc_input_type"]}2',
xlabel='UMAP1',
ylabel='UMAP2',
s=0.3,
alpha=0.5,
show_legend=True,
)The output UMAP plot will be saved at demo_output/analysis/umap_figures/UMAP_vqvec2.png by default.
# Compute bi-clustering heatmap
analysis.plot_clustermap(num_workers=4)
# Prepare image data
img = next(iter(datamanager.test_loader))['image'].detach().cpu().numpy()[:1]
# Compute index histogram
vqindhist1 = trainer.infer_embeddings(img, 'vqindhist1')
# Reorder the index histogram according to the bi-clustering heatmap
ft_spectrum = analysis.compute_feature_spectrum(vqindhist1)
# Generate a plot
import numpy as np
import matplotlib.pyplot as plt
x_max = ft_spectrum.shape[1] + 1
x_ticks = np.arange(0, x_max, 50)
fig, ax = plt.subplots(figsize=(10, 3))
ax.stairs(ft_spectrum[0], np.arange(x_max), fill=True)
ax.spines[['right', 'top']].set_visible(False)
ax.set_xlabel('Feature index')
ax.set_ylabel('Counts')
ax.set_xlim([0, x_max])
ax.set_xticks(x_ticks, analysis.feature_spectrum_indices[x_ticks])
fig.tight_layout()
fig.show()Rocky Linux 8.6, NVIDIA A100, CUDA 11.7 (GPU)
Ubuntu 20.04.3 LTS, NVIDIA 3090, CUDA 11.4 (GPU)
The full data used in this work can be found here.
The image data have the shape of [batch, 100, 100, 4], in which the last channel dimension corresponds to [target protein, nucleus, nuclear distance, nuclear segmentation].
Due to the large size, the whole data is split to 10 files. The files are intended to be concatenated together to form one large numpy file or one large csv.
Image_data00.npy
Image_data01.npy
Image_data02.npy
Image_data03.npy
Image_data04.npy
Image_data05.npy
Image_data06.npy
Image_data07.npy
Image_data08.npy
Image_data09.npy
Label_data00.csv
Label_data01.csv
Label_data02.csv
Label_data03.csv
Label_data04.csv
Label_data05.csv
Label_data06.csv
Label_data07.csv
Label_data08.csv
Label_data09.csv