De novo circular RNA detection from circular viral genome
- Bash (/bin/bash) in Linux or Mac OS
- Perl - https://www.perl.org
- BWA - http://bio-bwa.sourceforge.net (tested on 0.7.17)
- SAMtools - http://www.htslib.org (optional) to reduce disk usage by excluding unmapped reads from SAM file
- Perl module "GD" and "GD::Text::Align" - https://metacpan.org/pod/GD and https://metacpan.org/pod/GD::Text::Align (optional) to draw diagram
If you already have Git (https://git-scm.com) installed, you can get the latest development version using Git. It will take a few seconds.
git clone https://github.com/jiwoongbio/vircircRNA.git
./vircircRNA.sh <circular_chromosome.fasta> <gene.gff> <output.prefix> <input.1.fastq> [input.2.fastq]
- Demo (circular RNA detection from HPV genome). It will take a few minutes.
./vircircRNA.sh NC_001526.4.fasta NC_001526.4.gff3 SRS2410540 SRS2410540.part.1.fastq.gz SRS2410540.part.2.fastq.gz
- Generate concatenated sequence of circular chromosome
perl vircircRNA_chromosome.pl <circular_chromosome.fasta> > <concatenated_circular_chromosome.fasta>
- Mapping reads to concatenated sequence of circular chromosome
bwa index <concatenated_circular_chromosome.fasta>
bwa mem -T 19 -Y <concatenated_circular_chromosome.fasta> <input.1.fastq> [input.2.fastq] > <mapped_read.sam>
- Back-splice junction identification
perl vircircRNA_junction.pl -g <gene.gff> -A <junction.alignment.html> <mapped_read.sam> <concatenated_circular_chromosome.fasta> > <junction.txt>
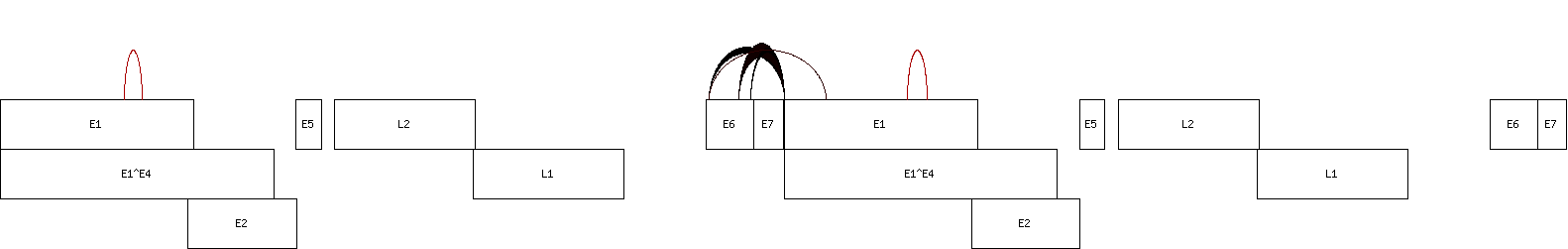
- Draw diagram
perl vircircRNA_diagram.pl -g <gene.gff> <junction.txt> <concatenated_circular_chromosome.fasta> <chromosome> > <diagram.png>
Zhao J, Lee EE, Kim J, Yang R, Chamseddin B, Ni C, Gusho E, Xie Y, Chiang CM, Buszczak M, Zhan X, Laimins L, Wang RC. "Transforming activity of an oncoprotein-encoding circular RNA from human papillomavirus" Nature Communications. 2019 May 24;10(1):2300. PMID: 31127091