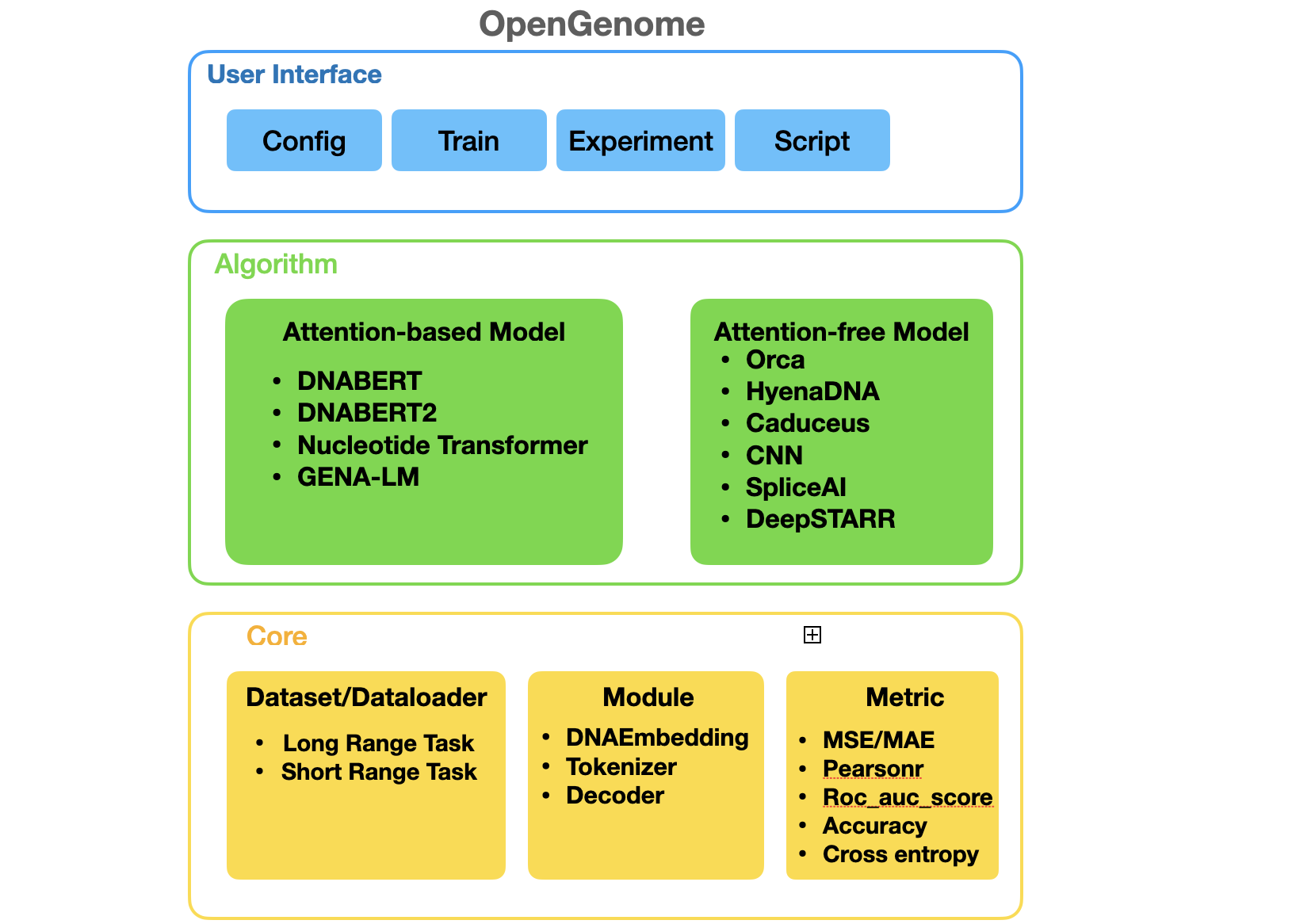
GenBench is a comprehensive benchmark for evaluating genomic foundation model, encompassing a broad spectrum of methods and diverse tasks, ranging from predicting gene location and function, identifying regulatory elements, and studying species evolution. GenBench offers a modular and extensible framework, excelling in user-friendliness, organization, and comprehensiveness. The codebase is organized into three abstracted layers, namely the core layer, algorithm layer, and user interface layer, arranged from the bottom to the top.
Code Structures
GenBench/configscontains configuration for benchmark evaluation.GenBench/datacontains datasets.GenBench/notebookcontains analysis and visualization notebooks.GenBench/srccontains source code for evaluation piplines.GenBench/weightcontains pretrained weights for benchmark evaluation.GenBench/experimentcontains scripts for experiment management.
This project has provided an environment setting file of conda, users can easily reproduce the environment by the following commands:
cd GenBench
conda env create -f environment.yml
conda activate OpenGenome
python setup.py developHere is an example of single GPU non-distributed training HyenaDNA on demo_human_or_worm dataset.
bash tools/prepare_data/download_mmnist.sh
python train.py -m train experiment=hg38/genomic_benchmark_mamba \
dataset.dataset_name=demo_human_or_worm \
wandb.id=demo_human_or_worm_hyenadna \
train.pretrained_model_path=path/to/pretrained_model \
trainer.devices=1Please see experiment.MD for the details of experiment management. and find scrips in 'experiment' directory
We support various Genomic foundation models. We are working on add new methods and collecting experiment results.
-
Spatiotemporal Prediction Methods.
-
Genomic foundation models Benchmarks.
Currently supported datasets
- Genomic benchmark (BMC Genomic Data'2023) [download] [config]
- GUE (Arxiv'2023) [download] [config]
- Promoter prediction (BioRxiv'2023) [download] [config]
- Splice site prediction (Cell Press'2019) [download] [config]
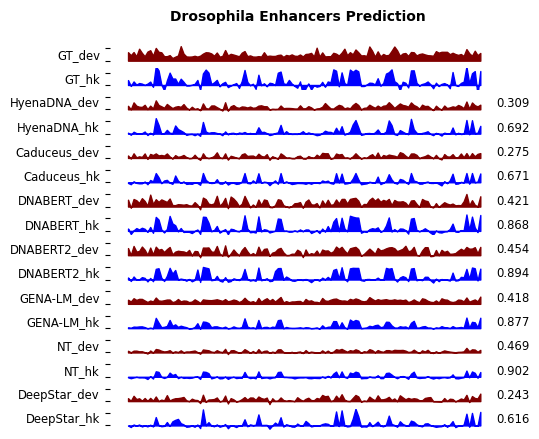
- Drosophila enhancer activity prediction (Nature Genetics'2022) [download] [config]
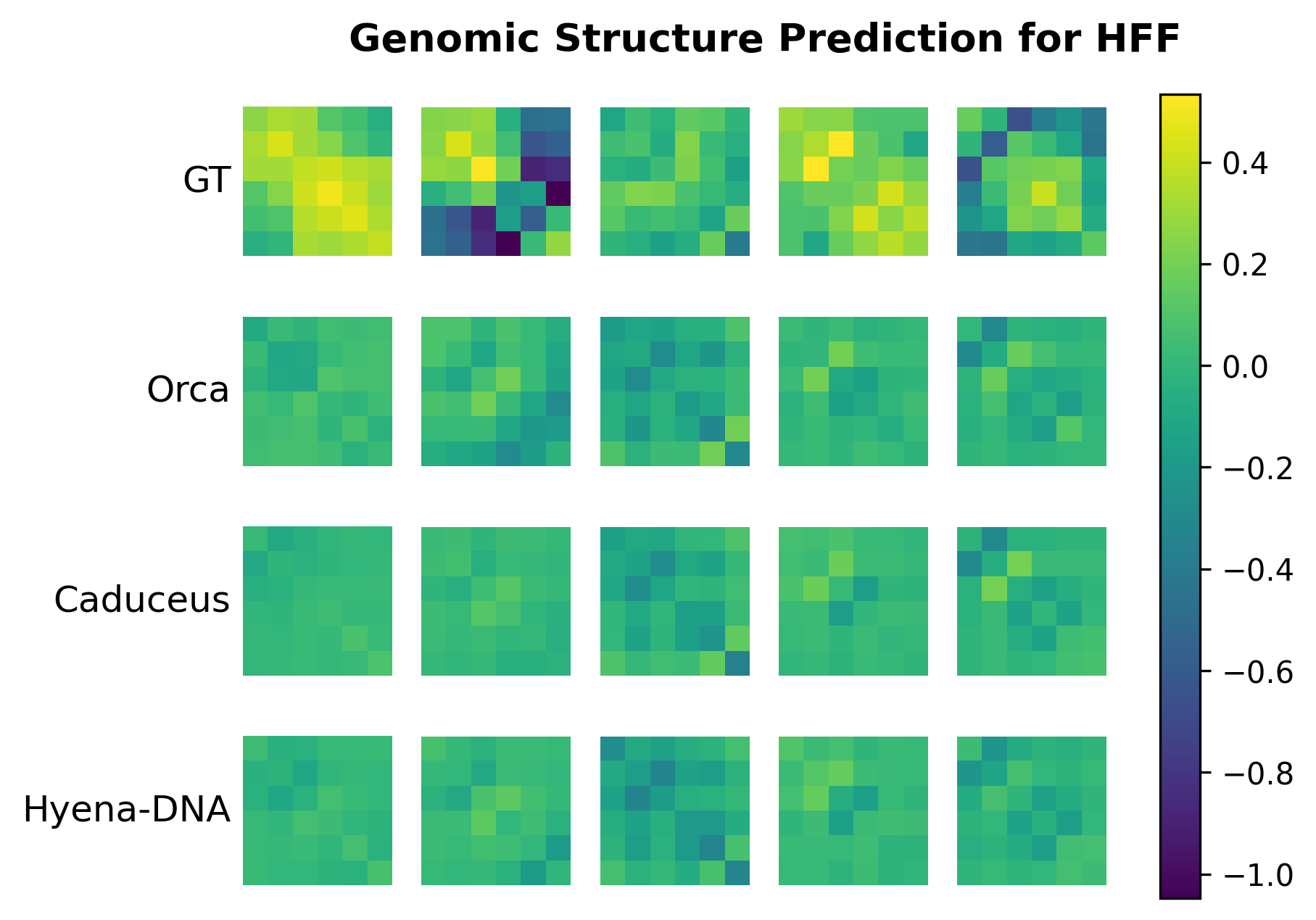
- Genomic Structure Prediction (Nature Genetics'2022) [download] [config]
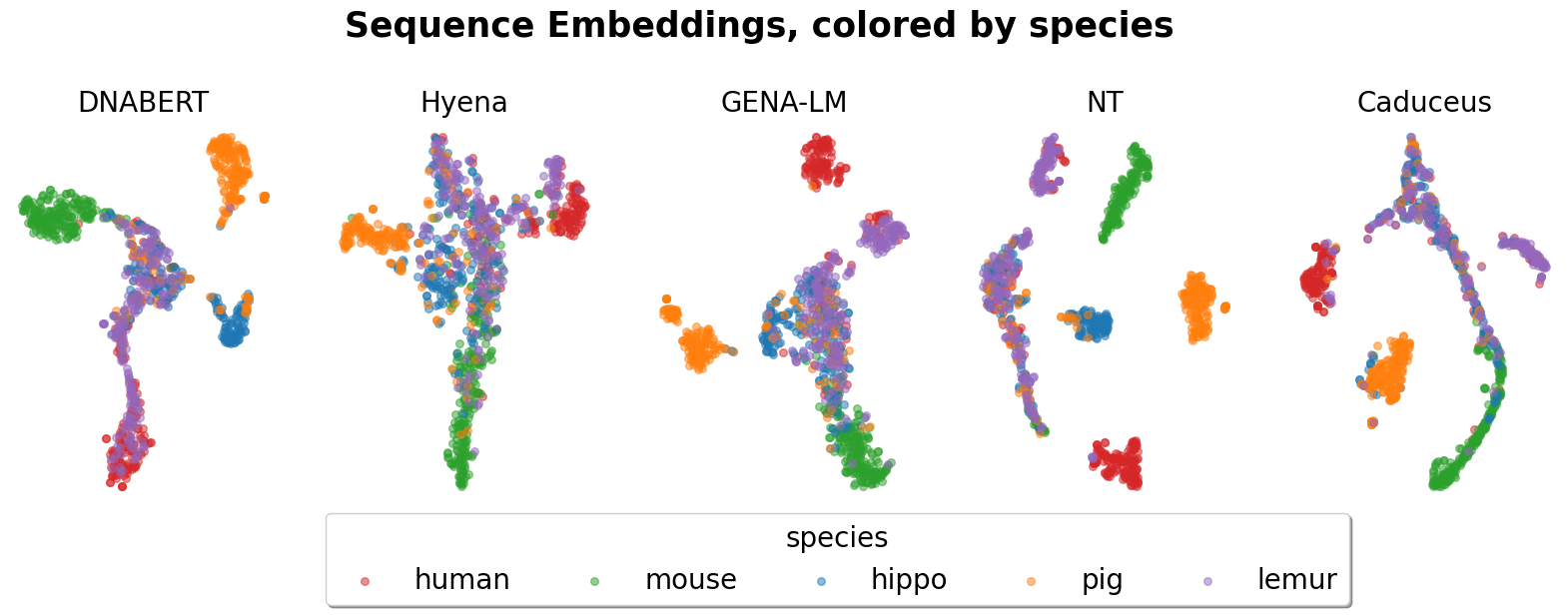
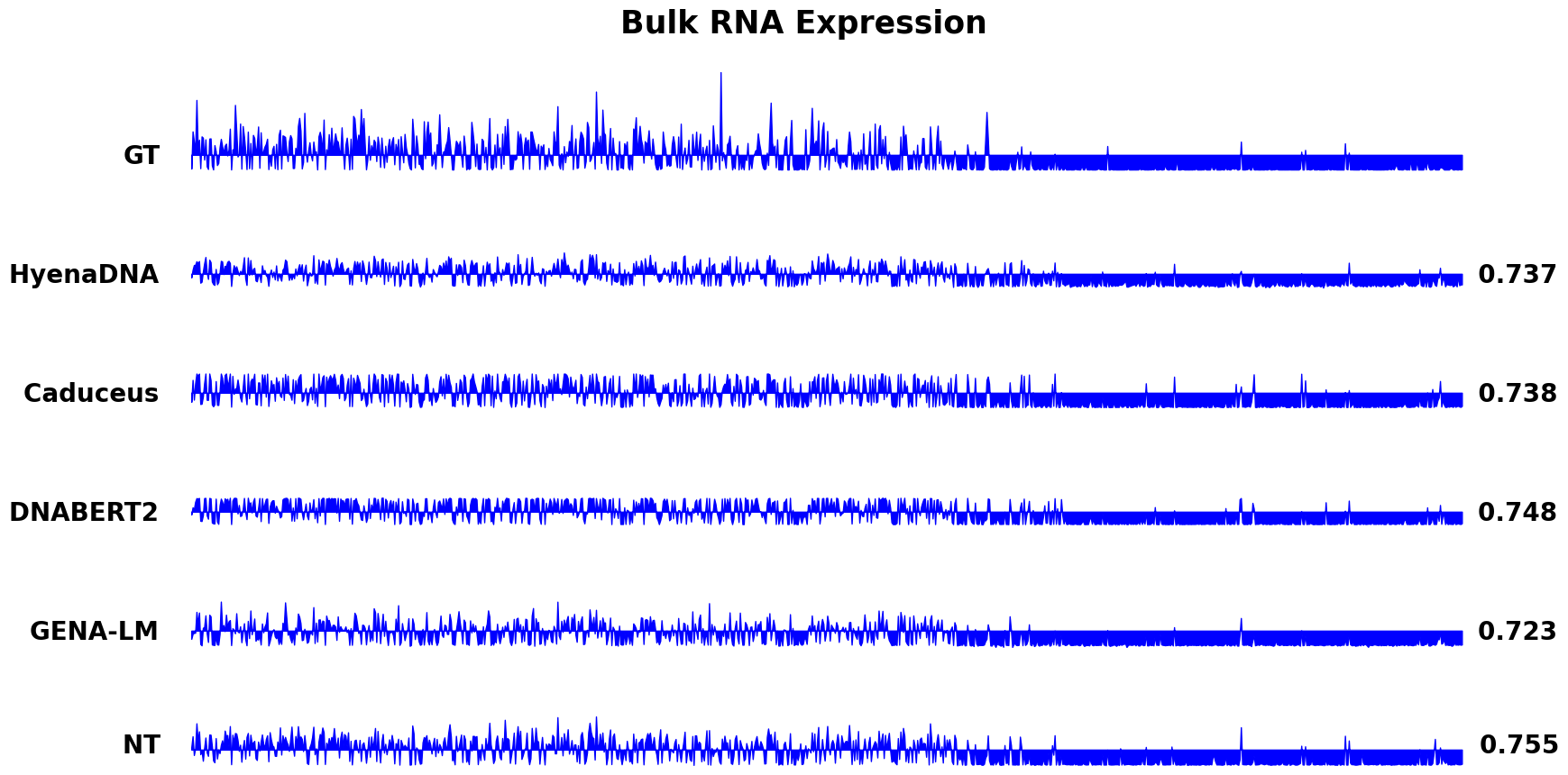
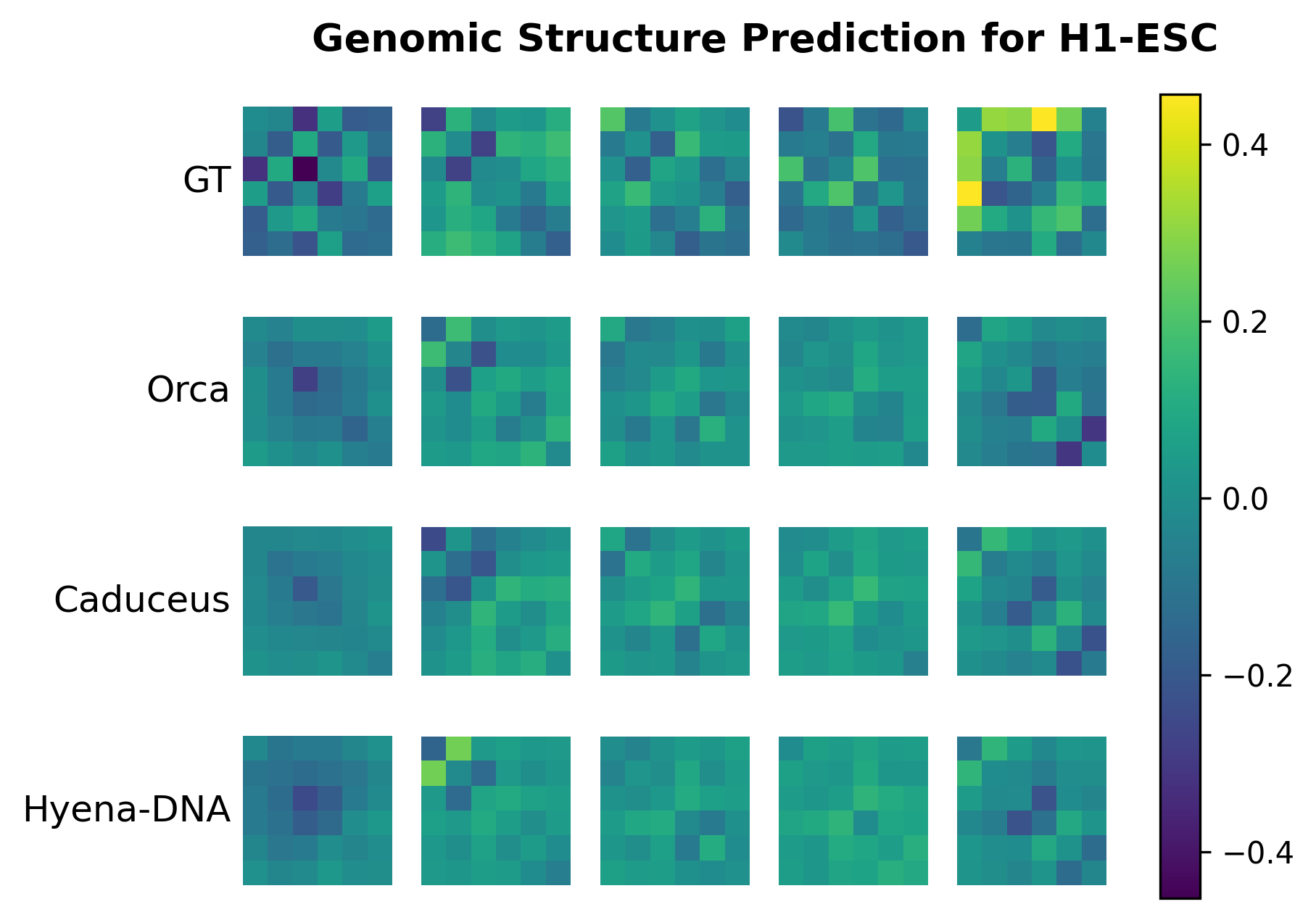
We present visualization examples of HyenaDNA below. For more detailed information, please refer to the notebook.
- For species classification task, visualization of t-sne embedding can be found in notebook/gene_cluster.ipynb.
- For visualization of Bulk RNA Expression, please refer to notebook/Bulk_prediction_spearman.ipynb.
- For Genomic Structure Prediction, visualization of predicted structures and ground truth structures are shown in notebook/plot_genomic_structure_h1esc.ipynb and notebook/plot_genomic_structure_hff.ipynb after running the experiment.
- for Drosophila enhancer activity prediction, visualization of predicted enhancers and ground truth enhancers are shown in notebook/drosophila_pearsonr.ipynb after running the experiment.
- for analysis of space complexity, please refer to notebook/count_flops.ipynb and for analysis of length effects and size effects, please refer to notebook/performance_length.ipynb and notebook/parameter_size.ipynb respectively.
This project is released under the Apache 2.0 license. See LICENSE for more information.
The framework of GenBench is insipred by HyenaDNA
- Jiahui Li(jiahuili.jimmy@gmail.com), Westlake University
- Zicheng Liu(liuzicheng@westlake.edu.cn), Westlake University