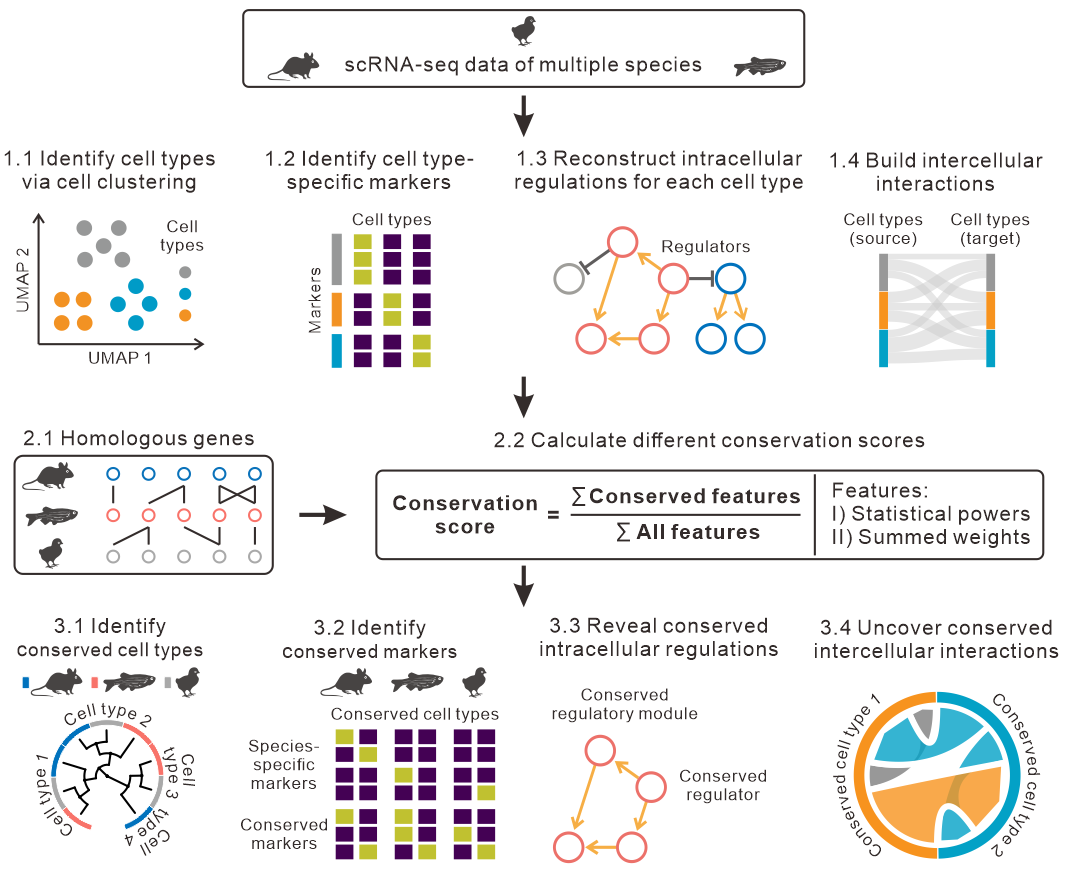
CACIMAR: cross-species analysis of cell identities, markers and regulations using single-cell sequencing profiles
CACIMAR has the following 4 main functions:
-
Identify all cell-type specific markers in each species
-
Identify species-specific or evolutionally conserved markers
-
Identify conserved intracellular regulations (gene regulatory networks)
-
Identify conserved intercellular interactions (cell-cell communications)
Install CACIMAR from github, run:
# install.packages("devtools")
devtools::install_github("jiang-junyao/CACIMAR")2024.08.02: Bugs in ‘Identify_CellType’ have been fixed.
For users who want to perform cross-species analysis quickly, we provide a brief tutorial based on small single-cell RNA sequencing (scRNA-seq) data to demonstrate the functionality and API of CACIMAR.
Quick tutorial for identifying evolutionally conserved celltype and markers based on scRNA-seq data
Quick tutorial for identifying evolutionally conserved gene regulatory networks
We also provide comprehensive examples to demonstrate the practical applications of CACIMAR.
Using CACIMAR to analyze retina scRNA-seq data from mouse and zebrafish
Using CACIMAR to analyze retina scRNA-seq data from mouse, zebrafish, and chick