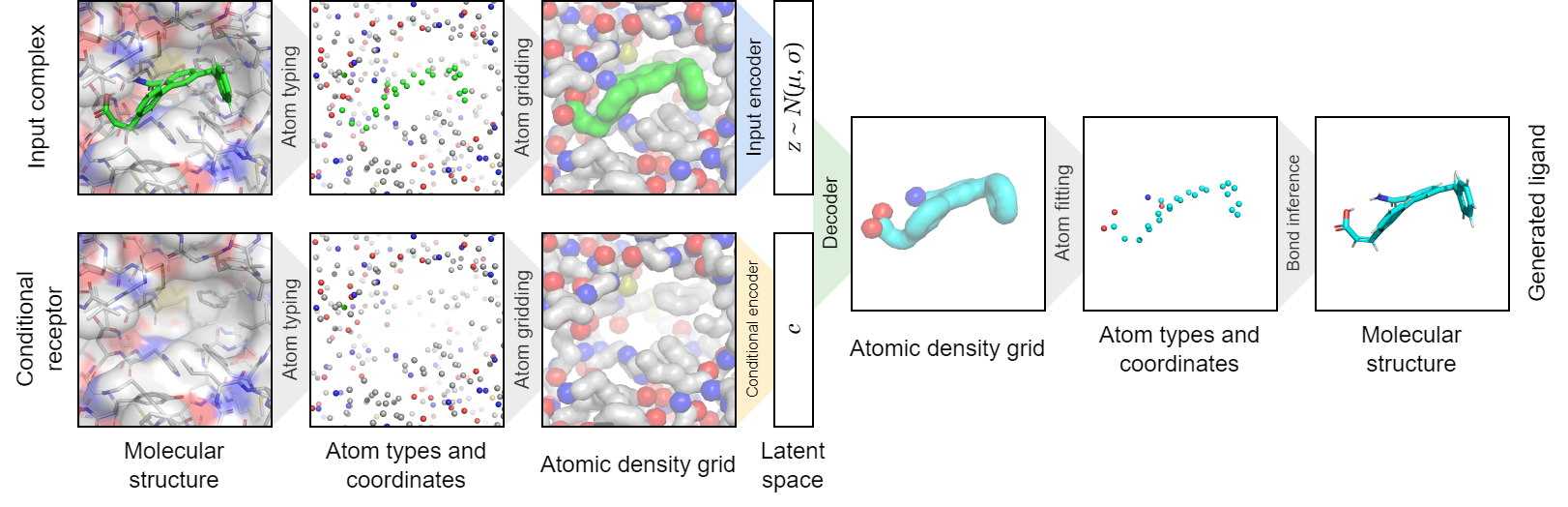
liGAN is a PyTorch project for structure-based drug discovery with deep generative models of atomic density grids.
NOTE: Please be aware that the current version of molgrid provided through pip/conda is incompatible with conda openbabel, and you will likely get segmentation faults if you install them both through conda.
A molgrid conda build recipe is in the works (see https://github.com/mattragoza/conda-molgrid), but for now you can use this environment to build libmolgrid from source.
To generate molecules, you must first download the pretrained model weights:
sh download_weights.sh
Then just run the generate.py script with the default configuration file:
python3 generate.py config/generate.config
To train a model from scratch, you must first download the full Crossdocked2020 data set:
sh download_data.sh
More info about this data set can be found here.
Then you can run the train.py script with the default configuration file:
python3 train.py config/train.config
@article{ragoza2022chemsci,
title={{Generating 3D molecules conditional on receptor binding sites with deep generative models}},
author={Matthew Ragoza and Tomohide Masuda and David Ryan Koes},
journal={Chem Sci},
month={Feb},
day={7},
year={2022},
volume={13},
pages={2701--2713},
doi={10.1039/D1SC05976A},
abstract={The goal of structure-based drug discovery is to find small molecules that bind to a given target protein. Deep learning has been used to generate drug-like molecules with certain cheminformatic properties, but has not yet been applied to generating 3D molecules predicted to bind to proteins by sampling the conditional distribution of protein–ligand binding interactions. In this work, we describe for the first time a deep learning system for generating 3D molecular structures conditioned on a receptor binding site. We approach the problem using a conditional variational autoencoder trained on an atomic density grid representation of cross-docked protein–ligand structures. We apply atom fitting and bond inference procedures to construct valid molecular conformations from generated atomic densities. We evaluate the properties of the generated molecules and demonstrate that they change significantly when conditioned on mutated receptors. We also explore the latent space learned by our generative model using sampling and interpolation techniques. This work opens the door for end-to-end prediction of stable bioactive molecules from protein structures with deep learning.},
}