The groupFusedMulti package provides methods for hierarchical variable
selection for models with covariate effects stratified by multiple
binary factors.
The development version can be installed using the devtools package:
devtools::install_github("jaredhuling/groupFusedMulti")or by cloning and building.
Load the groupFusedMulti package:
library(groupFusedMulti)Access help file for the main fitting function groupFusedMulti() by
running:
?groupFusedMultiHelp file for cross validation function cv.groupFusedMulti() can be
accessed by running:
?cv.groupFusedMultiSimulate multivariate response data where the responses have a natural grouping:
set.seed(123)
dat.sim <- gen_sparse_multivar_data(nvars = 10L,
noutcomes = 8L, ## 8-dim response vector
nobs = 100L,
num.nonzero.vars = 5,
outcome.groups = rbind(c(1,1,1,2,2,2,2,2),
c(1,1,1,2,2,3,3,3)))
# design matrices
x <- dat.sim$x
x.test <- dat.sim$x.test
# response vectors
y <- dat.sim$y
y.test <- dat.sim$y.test
# true data-generating coefficients
beta <- rbind(0,dat.sim$beta)
outcome_groups <- rbind(c(1,1,1,2,2,2,2,2),
c(1,1,1,2,2,3,3,3))First fit a model for a range of tuning parameter values (no cross validation perfomed here)
fit.gfm <- groupFusedMulti(x, y,
nlambda = 25,
lambda.fused = c(0.00001, 0.0001, 0.001),
outcome.groups = outcome_groups,
adaptive.lasso = TRUE, adaptive.fused = TRUE,
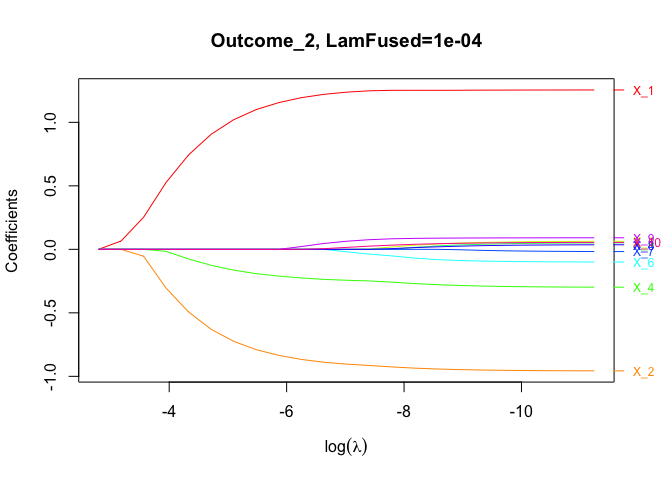
gamma = 0.5)Plot coefficient paths for the second response/outcome across all variables for the second fused lasso tuning parameter:
plot(fit.gfm, lam.fused.idx = 2,
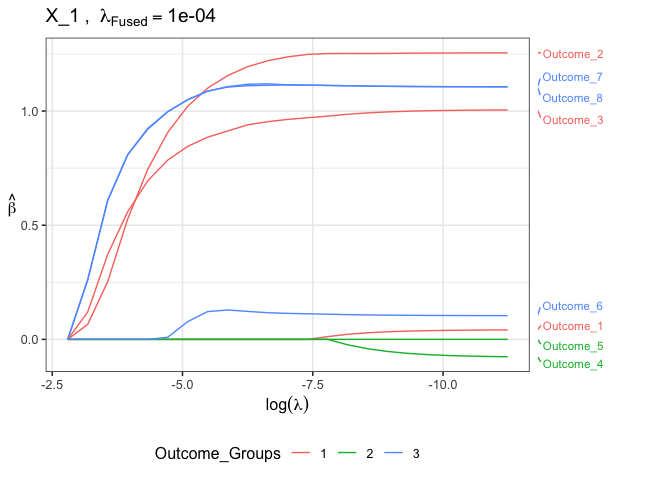
which.outcome = 2)Plot coefficient paths for a single variable across all outcomes for the second fused lasso tuning parameter:
plot(fit.gfm, lam.fused.idx = 2,
plot.type = "all_outcomes",
which.variable = 1, ## plot for first variable
which.outcome.group = 2) ## color according to second groupingFit groupFusedMulti model with tuning parameter selected with 5-fold
cross validation:
cvfit.gfm <- cv.groupFusedMulti(x, y,
nlambda = 50,
lambda.fused = c(0.000005, 0.00001, 0.000025, 0.00005, 0.0001),
outcome.groups = outcome_groups,
adaptive.lasso = TRUE, adaptive.fused = TRUE,
gamma = 0.5,
nfolds = 5)Compare estimated with true coefficients
est.coefs <- predict(cvfit.gfm, type = "coef")
colnames(beta) <- colnames(est.coefs)
rownames(beta) <- rownames(est.coefs)
round(est.coefs, 3)## Outcome_1 Outcome_2 Outcome_3 Outcome_4 Outcome_5 Outcome_6
## (Intercept) 0.047 -0.105 -0.087 -0.057 -0.057 -0.064
## X_1 0.000 1.146 0.908 0.000 0.000 0.131
## X_2 -0.440 -0.827 0.000 0.000 0.562 0.343
## X_3 0.000 0.000 0.000 0.000 0.000 0.016
## X_4 -0.312 -0.207 -0.381 0.000 0.000 -0.062
## X_5 -0.247 0.000 -0.327 0.000 0.000 -0.493
## X_6 0.000 0.000 0.000 -0.010 -0.001 -0.002
## X_7 0.000 0.000 0.000 0.000 0.014 0.052
## X_8 0.000 0.000 0.000 0.000 0.000 0.000
## X_9 0.000 0.000 0.000 0.000 0.000 0.000
## X_10 0.000 0.000 0.000 0.000 0.000 0.000
## Outcome_7 Outcome_8
## (Intercept) -0.102 -0.064
## X_1 1.103 1.103
## X_2 0.343 0.320
## X_3 -0.205 0.016
## X_4 -0.062 -0.022
## X_5 -0.148 -0.493
## X_6 0.000 0.000
## X_7 0.011 0.000
## X_8 0.000 0.000
## X_9 0.000 0.000
## X_10 0.000 0.000
beta## Outcome_1 Outcome_2 Outcome_3 Outcome_4 Outcome_5 Outcome_6
## (Intercept) 0.000 0.000 0.000 0.000 0.0 0.000
## X_1 0.000 1.000 1.000 0.000 0.0 0.000
## X_2 -0.500 -1.000 -0.125 0.000 0.5 0.500
## X_3 -0.125 -0.125 -0.125 0.000 0.0 0.000
## X_4 -0.125 -0.125 -0.125 -0.125 0.0 -0.125
## X_5 -0.500 0.000 -0.500 0.000 0.0 -0.500
## X_6 0.000 0.000 0.000 0.000 0.0 0.000
## X_7 0.000 0.000 0.000 0.000 0.0 0.000
## X_8 0.000 0.000 0.000 0.000 0.0 0.000
## X_9 0.000 0.000 0.000 0.000 0.0 0.000
## X_10 0.000 0.000 0.000 0.000 0.0 0.000
## Outcome_7 Outcome_8
## (Intercept) 0.000 0.000
## X_1 1.000 1.000
## X_2 0.500 0.500
## X_3 -0.500 0.125
## X_4 -0.125 -0.125
## X_5 0.000 -0.500
## X_6 0.000 0.000
## X_7 0.000 0.000
## X_8 0.000 0.000
## X_9 0.000 0.000
## X_10 0.000 0.000
Predict response for test data:
preds.gfm <- predict(cvfit.gfm, x.test, type = 'response')
str(preds.gfm)## num [1:100, 1:8] -0.993 -0.147 -0.171 0.113 0.848 ...
Evaluate mean squared error for each outcome:
sqrt(colMeans((y.test - preds.gfm) ^ 2))## [1] 0.9994061 1.1574990 1.0671242 1.0068098 0.9782761 1.0634526 1.1578628
## [8] 1.0385384
Average MSE across the 8 outcomes:
mean(sqrt(colMeans((y.test - preds.gfm) ^ 2)))## [1] 1.058621