SciPy John Hunter Excellence in Plotting Competition 2020
Submitted by Jan-Oliver Joswig, PhD student in the Molecular Dynamics Group at Freie Universität Berlin, Germany
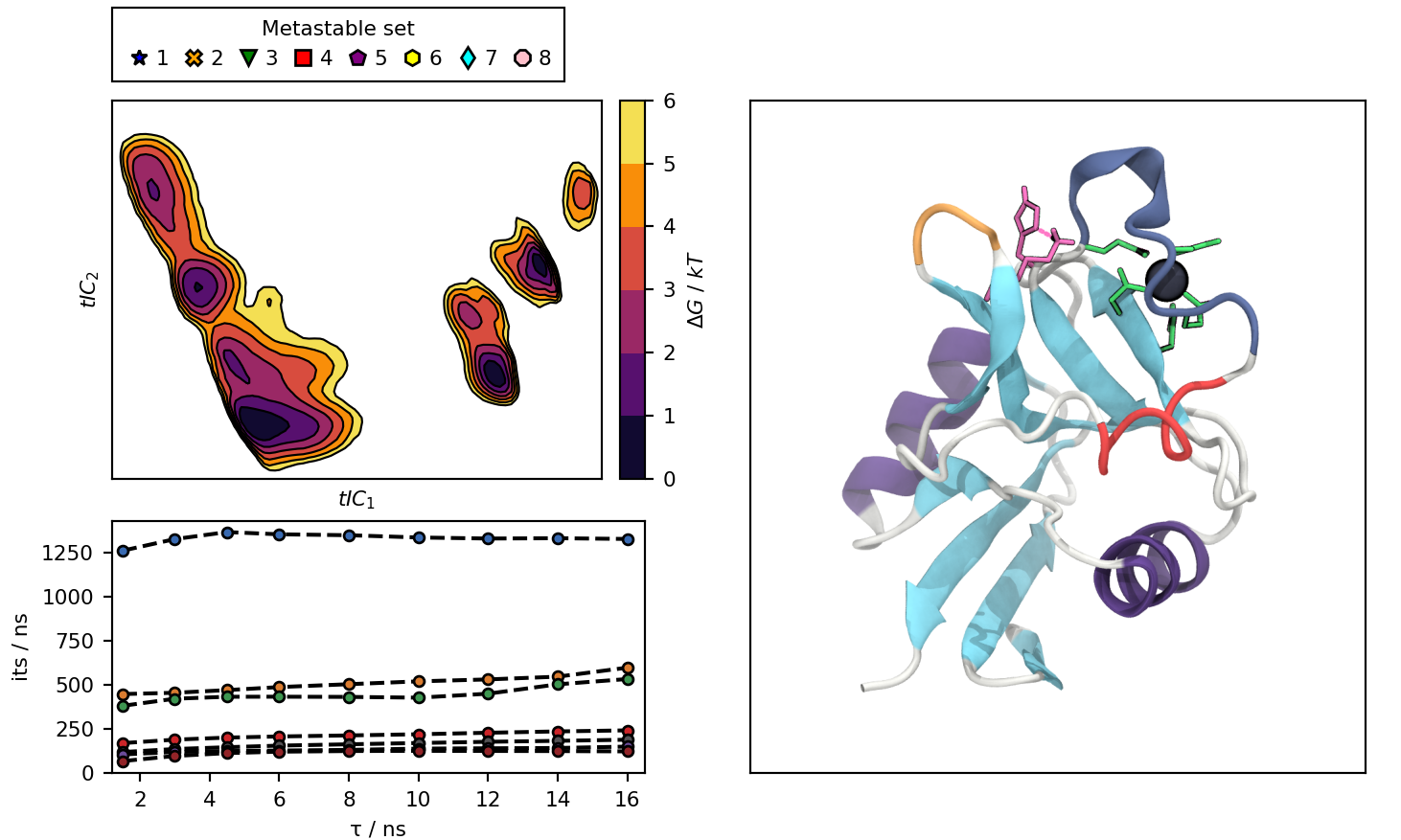
The notebook in this repository produces an interactive plot, with this default view:
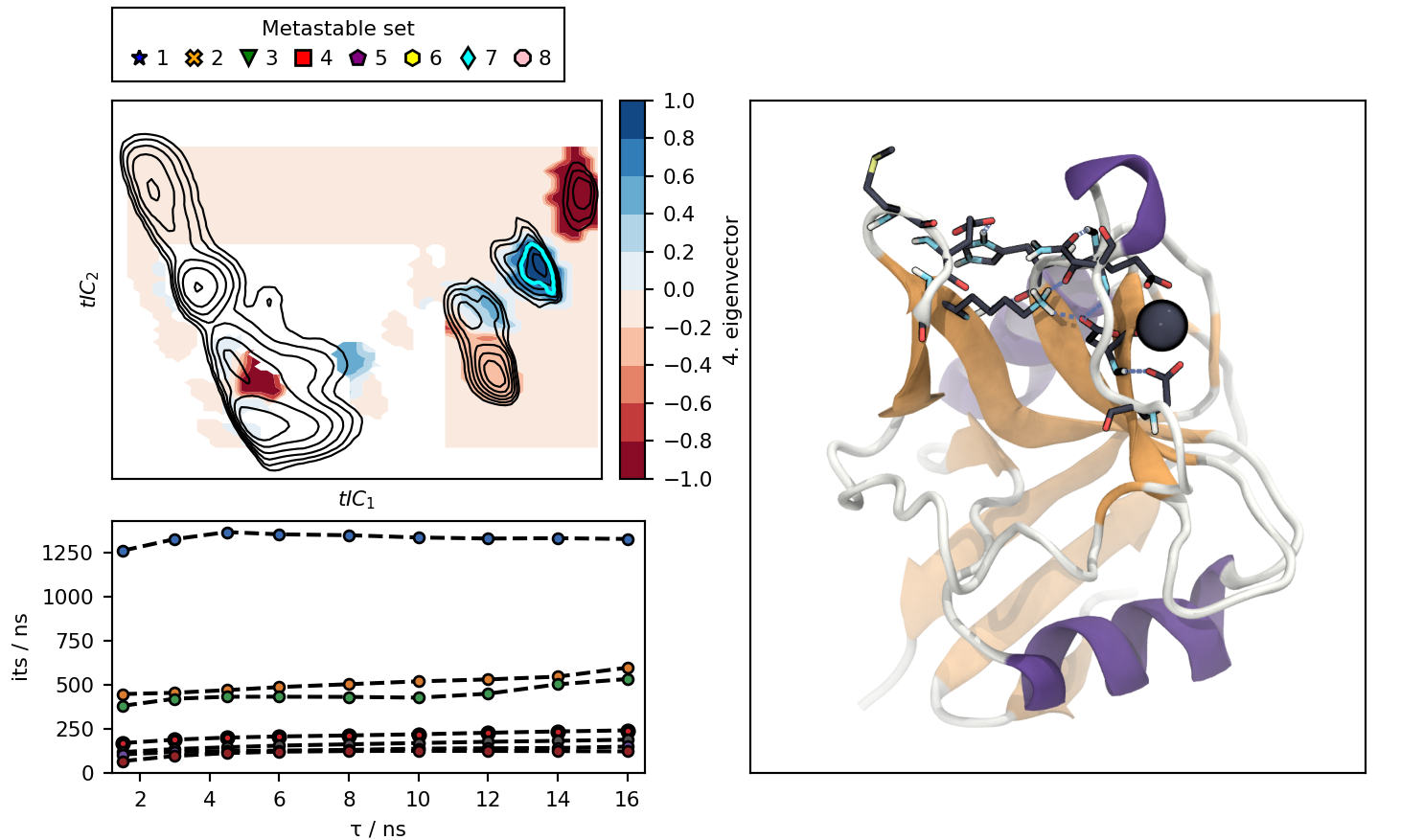
Clicking on the cluster icons in the legend or on the lines in the lower left plot, changes the illustration:
-
msm.ipynb : Jupyter notebook, containing a description of the background of this contribution, pre-requirements to draw the interactive plot, and the interactive plot itself.
-
data/ :
- dG.npy, x.npy, y.npy : free energy surface (histogram) and bin mids in two dimensions.
- dG_<cluster>.npy, x_<cluster>.npy, y_<cluster>.npy : free energy surfaces for clusters of conformations.
- E_<process>.npy, x_e<cluster>.npy, y_e<cluster>.npy : eigenvector surface for the transition processes.
- its.npy : implied time scales, corresponding to lag times of the MSMs.
- lags.npy : lag times for estimated MSMs.
- conf.png : image of langerin (default).
- cl<cluster>.png : image of a representative structure for the clusters.
-
default_view.png : Picture of the default state of the interactive plot.
-
cluster7_view.png : Picture of the interactive plot were cluster 7 and process 4 were picked.
-
screencast.mp4 : Video demonstrating the interactive plot.
-
matplotlibrc : Matplotlib plot configuration file to ensure a sensible layout in the notebook.