Learning Inter-Modal Correspondence and Phenotypes from Multi-Modal Electronic Health Records (TKDE)
This repo contains the PyTorch implementation of the paper Learning Inter-Modal Correspondence and Phenotypes from Multi-Modal Electronic Health Records in TKDE. [paper] [publisher]
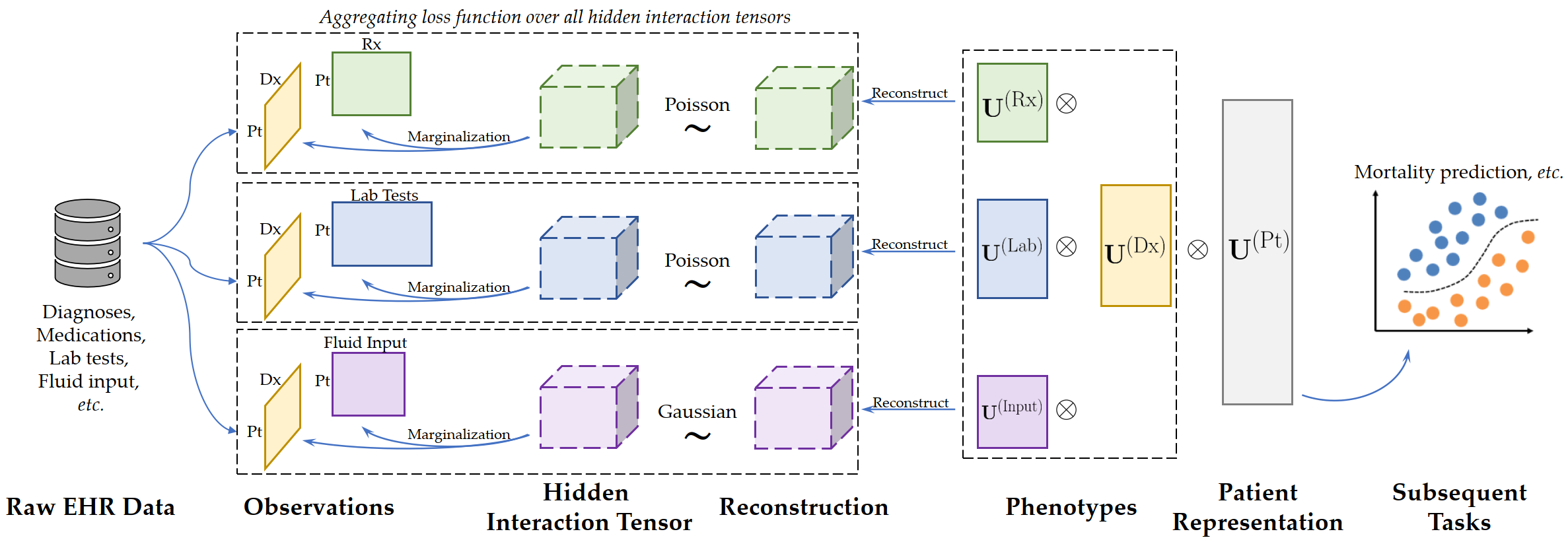
Overview: cHITF is a framework to jointly learn clinically meaningful phenotypes and hidden correspondence across different modalities from EHR data. Existing tensor factorization models mostly rely on the inter-modal correspondence to construct input tensors. In real-world datasets, the correspondence are often unknown and heuristics such as "equal-correspondence" are commonly used, leading to inevitable errors. cHITF aims to discover such unknown inter-modal correspondence simultaneously with the phenotypes.
Check out our paper for more details.
If you find the paper or the implementation helpful, please cite the following paper:
@article{yin2020chitf,
author = {Yin, Kejing and Cheung, William K. amd Fung, Benjamin C. M. and Poon, Jonathan},
title = {Learning Inter-Modal Correspondence and Phenotypes from Multi-Modal Electronic Health Records},
journal = "IEEE Transactions on Knowledge and Data Engineering (TKDE)",
year = "in press",
}This is an extension of our previous IJCAI-18 paper. You may also check out and cite the IJCAI version:
@inproceedings{yin2018joint,
title={Joint learning of phenotypes and diagnosis-medication correspondence via hidden interaction tensor factorization},
author={Yin, Kejing and Cheung, William K and Liu, Yang and Fung, Benjamin C. M. and Poon, Jonathan},
booktitle={Proceedings of the Twenty-Seventh International Joint Conference on Artificial Intelligence ({IJCAI-18})},
pages={3627--3633},
year={2018},
organization={AAAI Press}
}The codes have been tested with the following packages:
- Python 3.7
- PyTorch 1.3
- openpyxl 3.0.5
To run the model with a quick demo data, clone the repo and run the following commands:
git clone git@github.com:jakeykj/cHITF.git
cd cHITF
python main.pyThe results will be saved in the folder ./results/.
Use python main.py --help to obtain more information about setting the parameters of the model.
Specification of Hidden Interaction Tensors
The hidden interaction tensors can be specified using -M or --modalities argument. Each interaction tensor consists of two or more modalities which are joined with a hyphen -. Examples:
-M dx-rxruns cHITF with one single hidden interaction tensor of diagnosis and medications.-M dx-rx dx-labruns cHITF with two hidden interaction tensors: the first has diagnosis and medications and the second has diagnosis and lab tests.-M dx-rx-labruns cHITF with one single 4D hidden interaction tensor with diagnosis, medications, and lab tests. Although this is supported, it is NOT recommended to use hidden interaction tensors with more than two modalities due to much more complex interactions between them. See the paper for more discussions.
The distribution of each hidden interaction tensor must also be specified by using the -d or --distributions argument. We support two distributions: Poisson (letter: P) and Gaussian/Normal (letter: G). To specify the distribution, feed the letters as the argument in the same order as the hidden interaction tensor For example:
-M dx-rx -d Pspecifies to use Poisson distribution for the hidden interaction tensor.-M dx-rx dx-input -d P Gspecifies Poisson and Gaussian distributions for the first and second interaction tensors, respectively.
The data are organized in a Python Dict object and are saved using the Python built-in pickle module. The input matrices and the feature index-to-description mappings are stored in the Dict object:
dx: Thepatient-by-diagnosisbinary matrix in form ofnumpy.ndarray.rx: Thepatient-by-medicationcounting matrix in form ofnumpy.ndarray.lab: Thepatient-by-lab_testcounting matrix in form ofnumpy.ndarray.input: Thepatient-by-input_fuildreal-number matrix in form ofnumpy.ndarray.dx_idx2desc,rx_idx2desc,lab_idx2desc, andinput_idx2desc: Python Dict objects with the feature indices as keys and descriptions as values for modalities of diagnosis, medications, lab tests, and input fluids, respectively.
Note that matrices of all modalities must have the same size in their first dimensions.
The binary label of each patient is also one element of the Dict:
mortality: A Python List object containing the binary mortality label of each patient. Must have the same size as the first dimension of all matrices as above.
If you use other datasets, you can organize the input data in the same format described above, and pass the <DATA_PATH> as a parameter to the training script:
python main.py --input <DATA_PATH>If you have any enquires, please contact Mr. Kejing Yin by email:
cskjyin [AT] comp [DOT] hkbu.edu.hk. You can also raise issues in this repository.
👉 Check out my home page for more research work by us.