The RedoxPrediction project is the code for the paper
Predicting Redox Potentials by Graph-Based Machine Learning Methods (preprint).
- Python version 3.8 or higher.
python3 -m pip install -r requirements.txt
git clone https://github.com/jajupmochi/graphkit-learn
cd graphkit-learn
python setup.py [develop] [--user]
If you want to run the GNN models on GPUs, NVIDIA GPU and driver is required. Check this tutorial for more information.
git clone git@github.com:jajupmochi/RedoxPrediction.git
cd RedoxPrediction/
python setup.py [develop] [--user]
python3 redox_prediction/models/run_xps.py
It is also possible to perform a specific experiment by appending arguments, e.g.,
python3 redox_prediction/models/run_xps.py --dataset brem_togn_dGred --model nn:mpnn --descriptor atom_bond_types
The experiments can also be performed on a High Performance Computer cluster with SLURM installed, by simply running
python3 redox_prediction/jobs/run_job.py
The outputs will be stored in the redox_prediction/outputs/ directory.
The details of the dataset can be found in the following repository: ORedOx159.
Notice: in this project, the introduced dataset is also referred to by the name Redox or brem_togn.
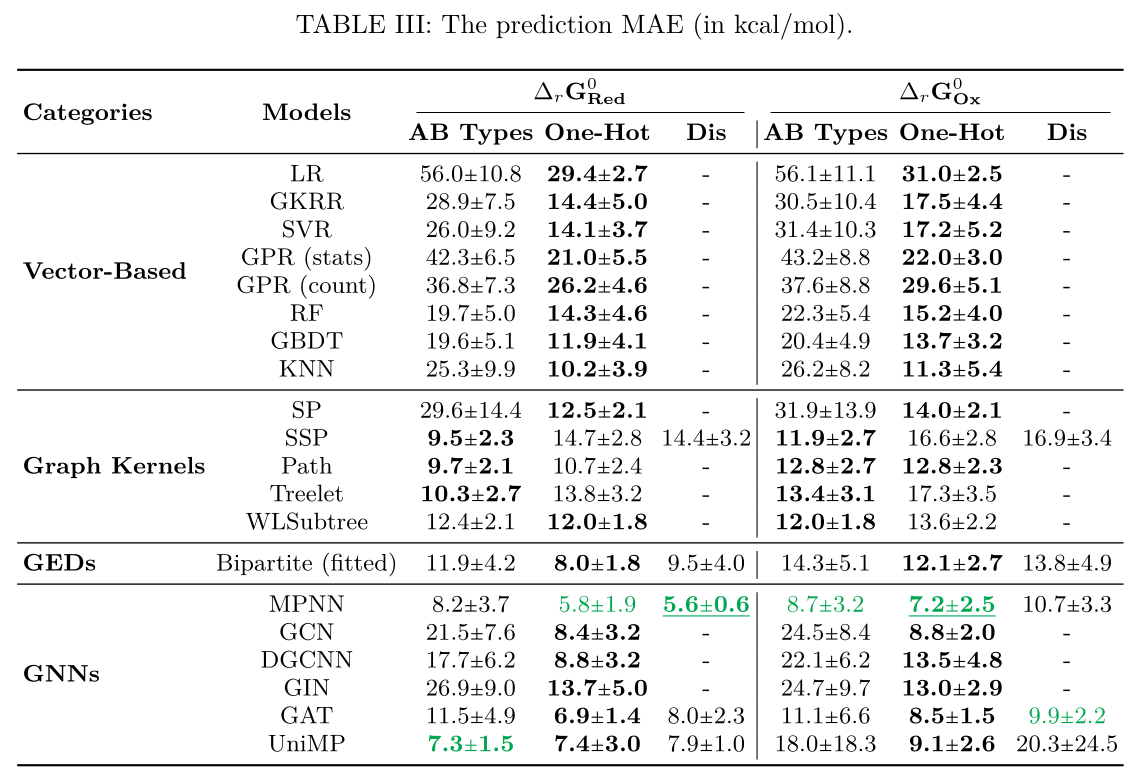
Results in terms of MAE, measured on the test sets. The "-" notation indicate that the method is not suitable for the descriptor:
Potentials computed by DFT vs the ones predicted via the best descriptor and model:
Reference time per datapoint for graph-based models, in log-scaled with base 10, and in seconds:
- Linlin Jia, the PRG group, Institute of Computer Science, University of Bern
- Éric Brémond, ITODYS, CNRS, Université Paris Cité
- Larissa Zaida, ITODYS, CNRS, Université Paris Cité
- Benoit Gaüzère, the LITIS Lab, INSA Rouen Normandie
- Vincent Tognetti, the COBRA Lab, Normandie Université
- Laurent Joubert, the COBRA Lab, Normandie Université
If you have used this library in your publication, please cite the following paper (coming soon):
E.B. and L.Z. thank Dr. P.P. Lainé (ITODYS lab, CNRS) for fruitful discussions regarding the construction of the ORedOx159 database.
E.B. gratefully acknowledges ANR (Agence Nationale de la Recherche) for the financial support of this work through the MoMoPlasm project (ANR-21-CE29-0003). He thanks also ANR and CGI (Commissariat à l’Investissement d’Avenir) for their financial support of this research through Labex SEAM (Science and Engineering for Advanced Materials and devices) ANR-10-LABX-096, ANR-18-IDEX-0001, and acknowledges TGGC (Très Grand Centre de Calcul du CEA) for computational resources allocation (Grant No. AD010810359R1).
This work has also been partially supported by University of Rouen Normandy, INSA Rouen Normandy, the “Centre National de la Recherche Scientifique” (CNRS), the European Regional Development Fund (ERDF), Labex SynOrg (ANR-11-LABX-0029), Carnot Institut I2C, the graduate school for research XL-Chem (ANR-18-EURE-0020 XL CHEM), and the “Région Normandie”. The Centre Régional Informatique et d’Applications Numériques de Normandie (CRIANN) is acknowledged for providing access to computational resources to L.J., V.T. and L.J.