This repository is an author's implementation of the paper DRACON: Disconnected Graph Neural Networkfor Atom Mapping in Chemical Reactions
We propose a model that affords both predicting reaction outcomes and finding atom mapping at the
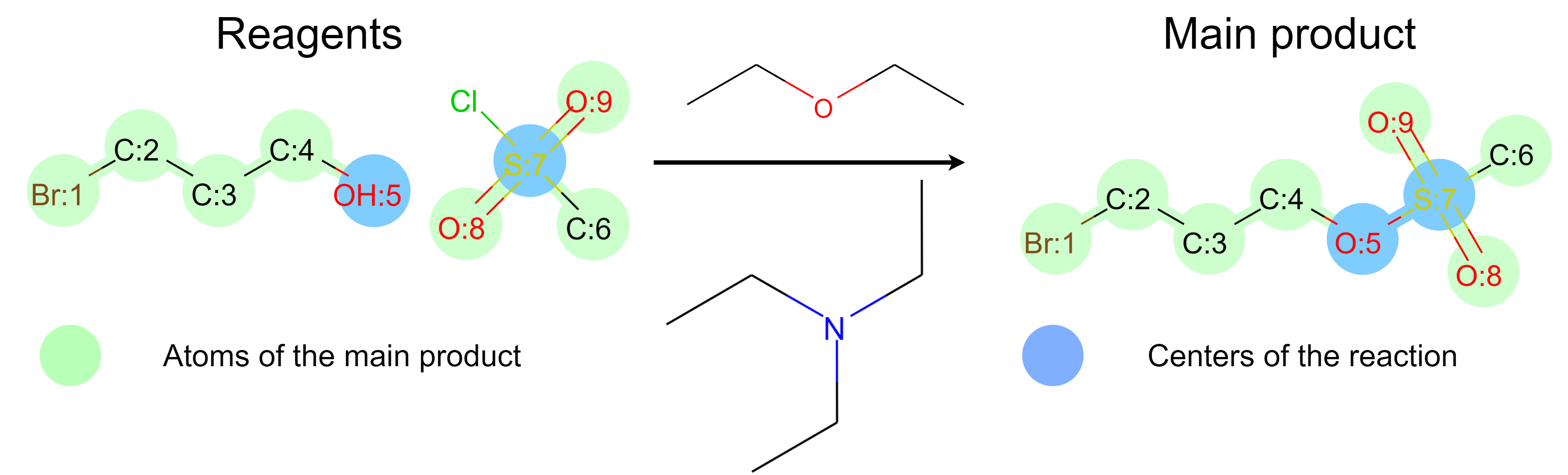
same time. Two specific tasks are solved in parallel (see Fig.~\ref{fig:reaction}). Atoms of the
main product and reaction centers are found. Centers of the reaction are atoms of the main product.
The atoms change their configuration during the reaction. The configuration of an atom is an
aggregate of characteristics of the atom and adjacent bonds. In terms of graph theory, both tasks
are node-classification in a disconnected graph of reactant and product molecules. The novel
Disconnected Graph Attention Convolution neural network
(DRACON) solves the node-classification tasks. Atoms of the main product and centers of the
reaction determine the outcome in the majority of reactions.
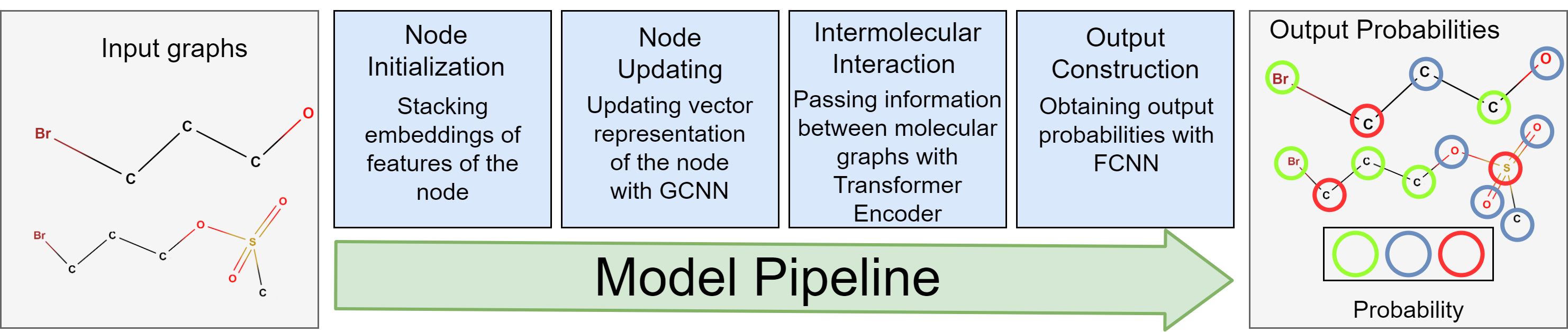
The overall model pipeline consists of four blocks. Firstly, each
atom is mapped to a real vector according to its characteristics in the molecule. The model uses
numerical characteristics of atoms. Secondly, the vectors are updated with Relational Graph
Convolution Neural Network (RGCNN). The RGCNN generalizes Graph
Convolution Neural Network for graphs with different edge types that correspond
to chemical bonds. In this work, we use extended molecular graphs with molecules' and reaction's
level nodes to pass information across different molecules. Then, the
Transformer encoder processes the vectors. The block simulates intermolecular interaction, which is
a mechanism of chemical reactions. Finally, the fully-connected neural network (FCNN) gives
probabilities for each atom in the node classification problems.
Compared with other works, DRACON has several novel aspects in terms of neural network architecture. DRACON generalizes the graph convolution neural network for the disconnected molecular graphs. The natural structure of the DRACON is suitable to add features of molecules, atoms like valencies, hybridization, types of chemical bonds, etc.
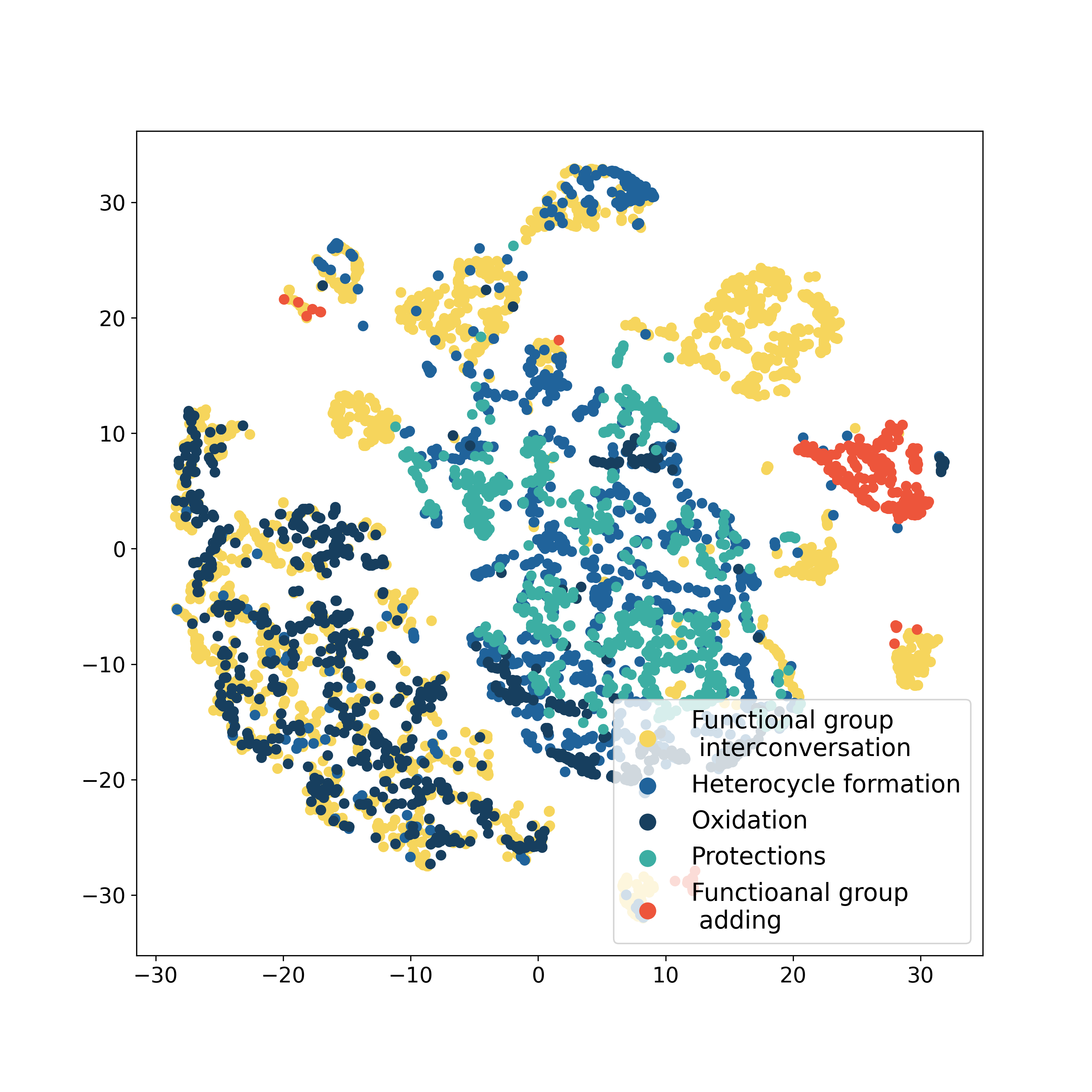
The authors investigate vector representations of reactions. The best model demonstrates that
pseudo-nodes in the extended graph of source molecules learn chemical information about the whole
reaction. Similar representation correspond chemical reactions which have a similar mechanism.
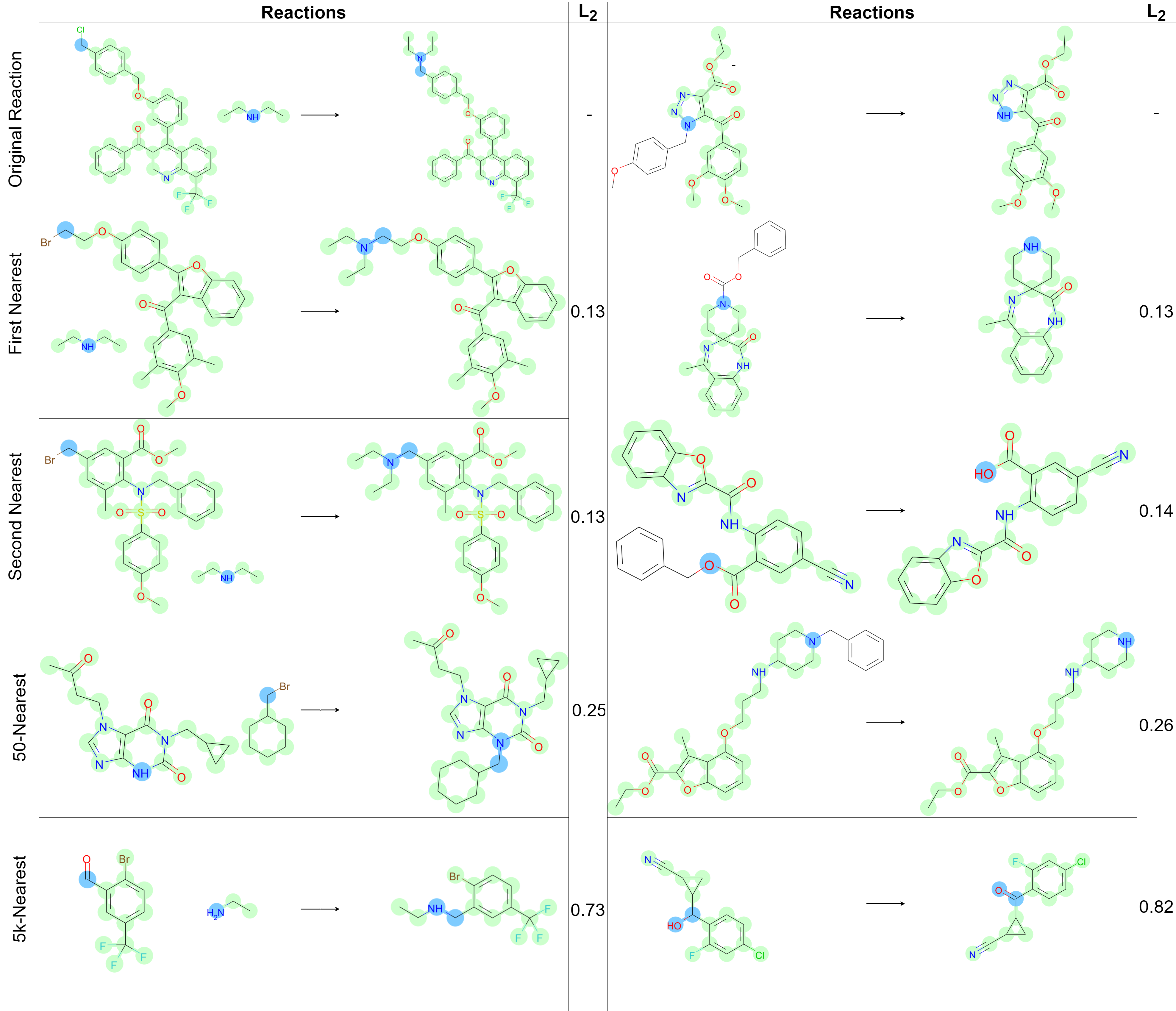
The model analysis illustrates that it gains substantial chemical insight, and one could differentiate and group chemical reactions by their types in a fully unsupervised fashion
Linux machine with Nvidia-1080Ti, Python 3.7.7 was used for the experiments with the folowing packages:
dgl-cu101==0.4
Django==3.0.7
matplotlib==3.2.1
numpy==1.18.2
pandas==1.0.3
PyYAML==5.3.1
scikit-learn
scipy
seaborn==0.10.0
torch==1.4.0
tqdm==4.45.0
rdkit
Our custom data preprocessor takes pandas dataframes stored in pickle. The dataframes should consist column 'smarts' with canonical reactions, and different other columns which will be stored in the processed dataset.
To create dataset, you should use the following commands:
cd scripts
python -u process_dataset.py --raw_dataset 'path to raw pickles directory' --save_path 'save directory'
To train a model, you should use the following commands:
cd scripts
python -u experiment.py --config path to YAML configuration --device name of using device
Examples of .yml configurations are in experiments directory.
We created an online demo which source code is in server directory. The demo is available at http://93.175.29.159:8000/.