QC pipeline for SARS-CoV-2 sequencing and assembly data submitted to the ENA.
Provides basic QC information on where in the reference genome there is/not coverage. Provides masks of a consensus assembly, showing which bases are supported by the majority of reads (the proportion is a parameter)
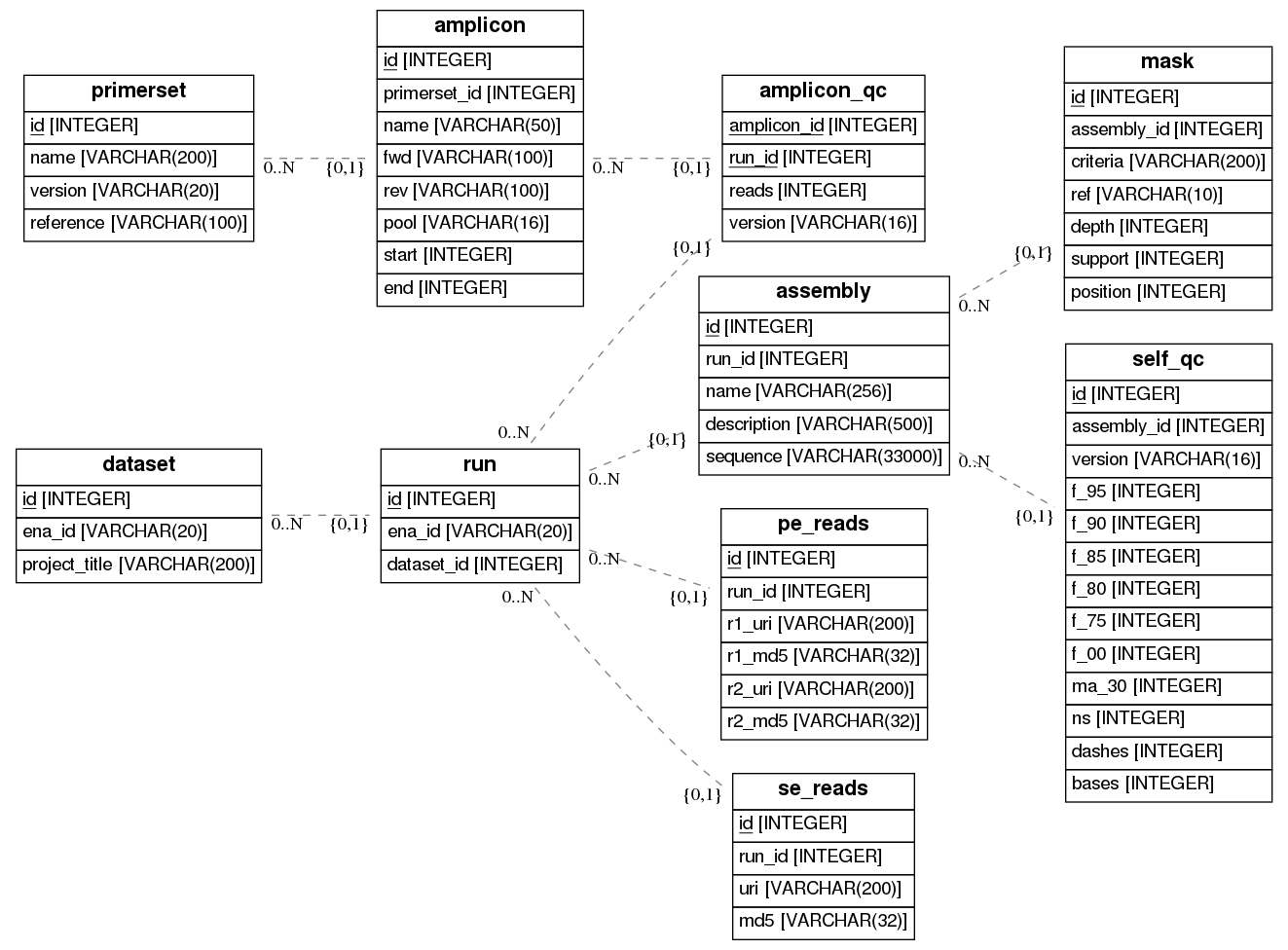
The processing therefore splits into two streams
- Per sample, map reads to the reference and mark positions which do not have read support If the user has provided information on the primer set used, this is used to infer which amplicons have dropped out. If the user has not provided primer information, generic pseudo-primer windows are tiles across the genome, and we measure which of those have been dropped.
- Per consensus assembly, map reads to the consensus and mask positions which do not have read support
All data written to a SQL database, including the masks.
This repository includes tools for managing a database of these QC results.
See the output/spec.md file
For complete details see qcovid.py --help
QCovid uses the SQLalchemy ORM. In general, qcovid.py is passed a database connection string as the first argument.
To initialise a local sqlite instance:
qcovid.py sqlite:///my_db.sqlite init
This will create an instance of the QCovid database in file named my_db.sqlite in the current working directory.
You may see a brief summary of the database contents with:
qcovid.py sqlite:///my_db.sqlite info
Initialise a project:
qcovid.py sqlite:///my_db.sqlite project PRJNAxxxxxx --title "project description"
This pipeline is meant to be run on data which has been accessioned in the ENA. Fetch target runs by project use enaDataBrowser:
enaGroupGet.py PRJNAxxxxxx -w -f fastq -d projects/
This will populate a directory for the project and subdirectories for each run's fastq files.
The qcovid.py tool can be used to import these samples into the database:
qcovid.py sqlite:///my_db.sqlite load PRJNAxxxxxx --dir Downloads/
This function will crawl through the downloaded project directory and populate the PairedRead and SingleRead tables.
For the assembly QC, import fasta file:
qcovid.py sqlite:///my_db.sqlite load --assemblies Downloads/fastas PRJNAxxxxxx
QCovid will infer the sample name from the fasta file name.
A fasta including all assemblies for a sample can be dumped:
qcovid.py sqlite:///my_db.sqlite fasta ERRxxxxxx
qcovid.py sqlite:///my_db.sqlite run will invoke the pipeline for samples which have fastq files but no analysis output.
This QC pipeline considers raw sequencing data and is flexible over:
- Single vs. paired reads (i.e. Nanopore vs. Illumina)
- Amplicon primers
We use bwa to map reads as part of the pipeline. Downstream analysis of the mapping is agnostic to mate pairing.
Primers should be presented as a list of tab delimited names, start positions, and end positions.
e.g. a bed file:
#Reference: MN908947.fasta (29903 bp.)
SARS-CoV-2_1_pool1 30 495
SARS-CoV-2_3_pool1 704 1205
SARS-CoV-2_5_pool1 1372 1897
SARS-CoV-2_7_pool1 2115 2642
SARS-CoV-2_9_pool1 2786 3257
SARS-CoV-2_11_pool1 3460 3948
SARS-CoV-2_13_pool1 4111 4634
SARS-CoV-2_15_pool1 4809 5359
SARS-CoV-2_17_pool1 5563 6037
Import a primer set into the database using the qcovid.py utility:
qcovid.py import primers.bed
There are about a dozen primer sets. Most samples post-April 2020 are ARTIC-v3.
Primer sets are not defined consistently and have to be preprocessed into these bed files individually. In general this entails parsing some list of sequences and putting them into a consistent coordinate system. We are using MN908947. If possible the amplicon names should follow the convention SARS-CoV-2_{number}_{pool} where number is the ID of the amplicon and pool specifies which (if any) reaction mix the primer is included in.
For reference these scripts and raw primer sets are included in primers/.
More information can be found in primers/README.md
For single-end reads and the Dutch primers (PRJEB38388), one would run:
bwa mem MN908947.fasta $PROJECT/$SAMPLE_1.fastq.gz | samtools view -bS - > $WORK/$SAMPLE.MN908947.bam
samtools sort $WORK/$SAMPLE.MN908947.bam -o $WORK/$SAMPLE.MN908947.sorted.bam
samtools index $WORK/$SAMPLE.MN908947.sorted.bam
python3 bin_amplicons.py primers/nCoV-nl-primal500-75.bed $SAMPLE.MN908947.sorted.bam
samtools sort $WORK/$SAMPLE.amplicons.bam -o $WORK/$SAMPLE.amplicons.sorted.bam
samtools index $WORK/$SAMPLE.amplicons.sorted.bamSee QCovid_plots.ipynb for examples.
bin_amplicons.py will optionally write reads that positively identify as amplicons to a 'cleaned' sam file.