See the pkdown site here.
Biological studies, especially in ecology, health sciences and taxonomy, need to describe the biological composition of samples. During the last twenty years, (i) the development of DNA sequencing, (ii) reference databases, (iii) high-throughput sequencing (HTS), and (iv) bioinformatics resources have allowed the description of biological communities through metabarcoding. Metabarcoding involves the sequencing of millions (meta-) of short regions of specific DNA (-barcoding, Valentini, Pompanon, and Taberlet (2009)) often from environmental samples (eDNA, Taberlet et al. (2012)) such as human stomach contents, lake water, soil and air.
MiscMetabar aims to facilitate the description,
transformation, exploration and reproducibility of
metabarcoding analysis using R. The development of MiscMetabar relies
heavily on the R packages
dada2,
phyloseq and
targets.
There is no CRAN version of MiscMetabar for now (work in progress). As MiscMetabar heavily relies on two bioconductor packages (dada and phyloseq), we need to first install those 2 packages using BiocManager.
You can install the stable version from GitHub with:
if (!require("BiocManager", quietly = TRUE)){
install.packages("BiocManager")
}
BiocManager::install("dada2")
BiocManager::install("phyloseq")
if (!require("devtools", quietly = TRUE)){
install.packages("devtools")
}
devtools::install_github("adrientaudiere/MiscMetabar")You can install the developement version from GitHub with:
if (!require("BiocManager", quietly = TRUE)) {
install.packages("BiocManager")
}
BiocManager::install("dada2")
BiocManager::install("phyloseq")
if (!require("devtools", quietly = TRUE)){
install.packages("devtools")
}
devtools::install_github("adrientaudiere/MiscMetabar", ref = "dev")See vignettes in the MiscMetabar website for more examples.
library("MiscMetabar")
library("phyloseq")
library("magrittr")
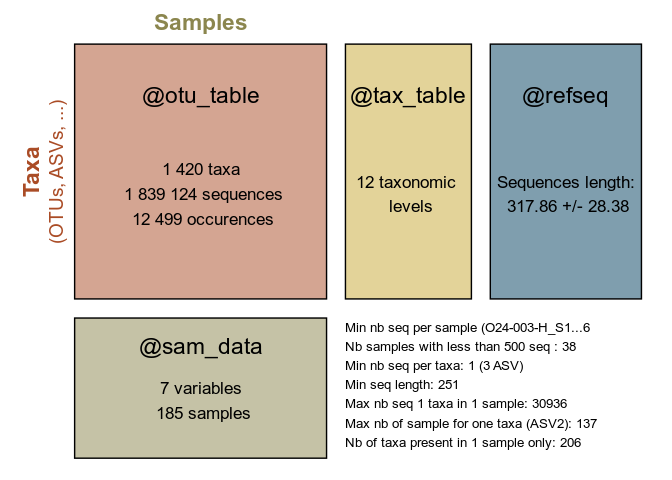
data("data_fungi")
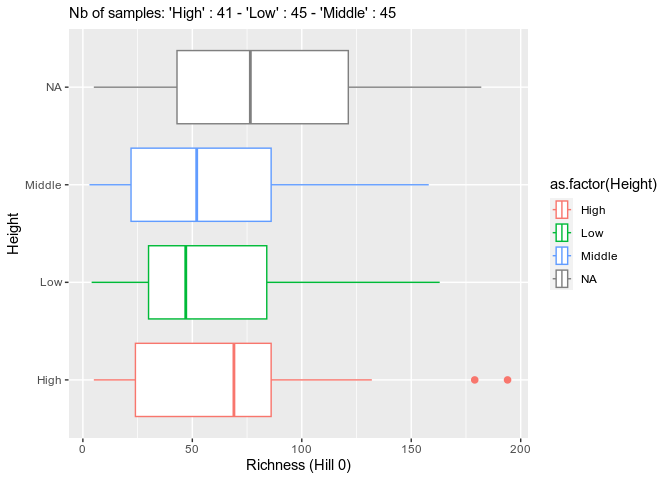
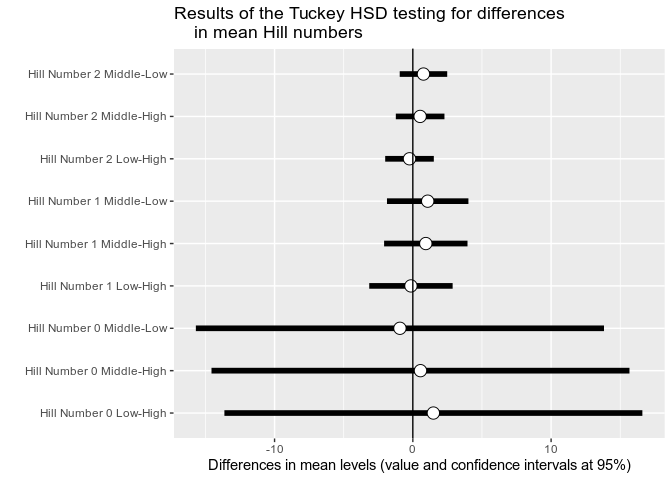
summary_plot_pq(data_fungi)p <- MiscMetabar::hill_pq(data_fungi, variable = "Height")
#> Taxa are now in rows.
#> Cleaning suppress 0 taxa and 0 samples.
p$plot_Hill_0p$plot_tuckeyif (!require("ggVennDiagram", quietly = TRUE)){
install.packages("ggVennDiagram")
}
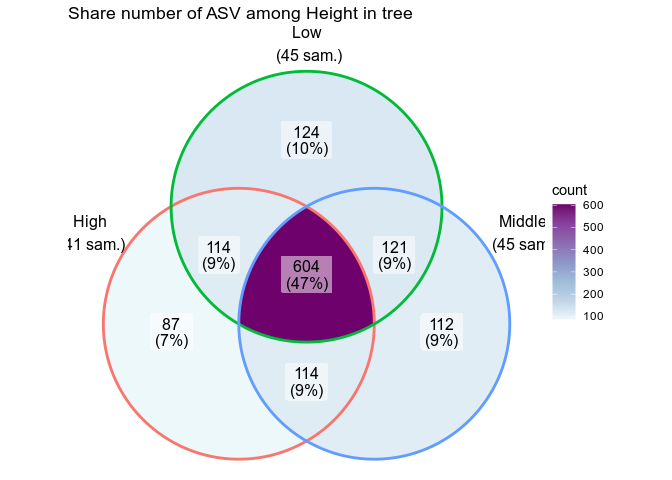
ggvenn_pq(data_fungi, fact = "Height") +
ggplot2::scale_fill_distiller(palette = "BuPu", direction = 1) +
labs(title = "Share number of ASV among Height in tree")sudo apt-get install ncbi-blast+sudo apt-get install vsearchTaberlet, Pierre, Eric Coissac, Mehrdad Hajibabaei, and Loren H Rieseberg. 2012. “Environmental Dna.” Molecular Ecology. Wiley Online Library. https://doi.org/10.1002/(issn)2637-4943.
Valentini, Alice, François Pompanon, and Pierre Taberlet. 2009. “DNA Barcoding for Ecologists.” Trends in Ecology & Evolution 24 (2): 110–17. https://doi.org/10.1016/j.tree.2008.09.011.