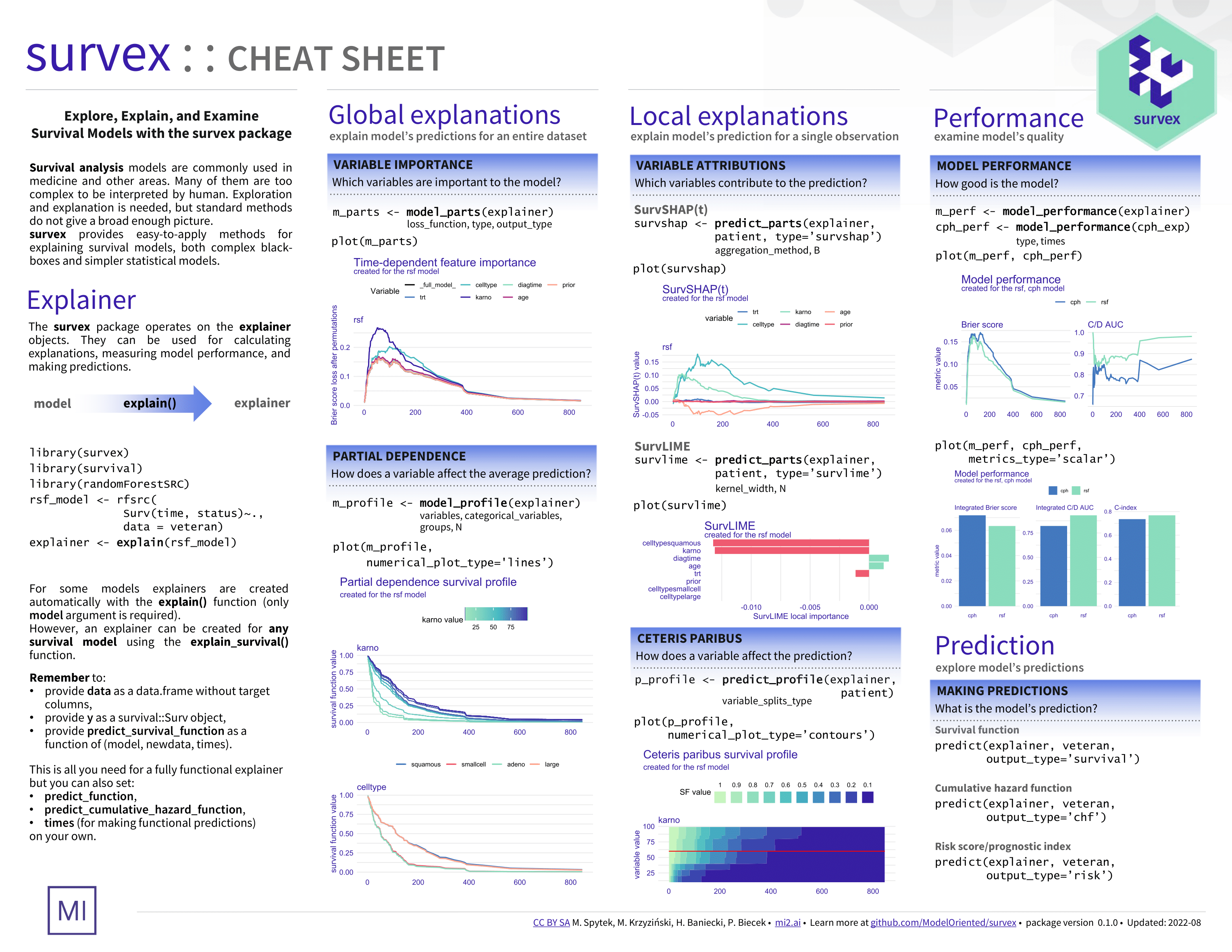
Survival analysis is a task dealing with time-to-event prediction. Aside from the well-understood models like CPH, many more complex models have recently emerged, but most lack interpretability. Due to a functional type of prediction, either in the form of survival function or cumulative hazard function, standard model-agnostic explanations cannot be applied directly.
The survex package provides model-agnostic explanations for machine learning survival models. It is based on the DALEX package. If you're unfamiliar with explainable machine learning, consider referring to the Explanatory Model Analysis book -- most of the methods included in survex extend these described in EMA and implemented in DALEX but to models with functional output.
The main explain() function uses a model and data to create a standardized explainer object, which is further used as an interface for calculating predictions. We automate creating explainers from the following packages: mlr3proba, censored, ranger, randomForestSRC, and survival. Raise an Issue on GitHub if you find models from other packages that we can incorporate into the explain() interface.
Note that an explainer can be created for any survival model, using the explain_survival() function by passing model, data, y, and predict_survival_function arguments.
The package is available on CRAN:
install.packages("survex")The latest development version can be installed from GitHub using devtools::install_github():
devtools::install_github("https://github.com/ModelOriented/survex")library("survex")
library("survival")
library("ranger")
# create a model
model <- ranger(Surv(time, status) ~ ., data = veteran)
# create an explainer
explainer <- explain(model,
data = veteran[, -c(3, 4)],
y = Surv(veteran$time, veteran$status))
# evaluate the model
model_performance(explainer)
# visualize permutation-based feature importance
plot(model_parts(explainer))
# explain one prediction with SurvSHAP(t)
plot(predict_parts(explainer, veteran[1, -c(3, 4)]))If you use survex, please cite it as
M. Spytek, M. Krzyziński, H. Baniecki, P. Biecek. survex: Explainable Machine Learning in Survival Analysis. R package version 0.2.2, 2022. https://github.com/ModelOriented/survex
@article{spytek2022survex,
title = {{survex: Explainable Machine Learning in Survival Analysis}},
author = {Mikołaj Spytek and Mateusz Krzyziński and
Hubert Baniecki and Przemysław Biecek},
journal = {R package version 0.2.2},
year = {2022},
url = {https://github.com/ModelOriented/survex}
}
- H. Baniecki, B. Sobieski, P. Bombiński, P. Szatkowski, P. Biecek. Hospital Length of Stay Prediction Based on Multi-modal Data towards Trustworthy Human-AI Collaboration in Radiomics. International Conference on Artificial Intelligence in Medicine, 2023.
- Share it with us!
- H. Ishwaran, U. B. Kogalur, E. H. Blackstone, M. S. Lauer. Random survival forests. Annals of Applied Statistics, 2008.
- A. Grudziąż, A. Gosiewska, P. Biecek. survxai: an R package for structure-agnostic explanations of survival models. Journal of Open Source Software, 2018.
- M. S. Kovalev, L. V. Utkin, E. M. Kasimov. SurvLIME: A method for explaining machine learning survival models. Knowledge-Based Systems, 2020.
- R. Sonabend, F. J. Király, A. Bender, B. Bischl, M. Lang. mlr3proba: an R package for machine learning in survival analysis. Bioinformatics, 2021.
- E. Hvitfeldt, H. Frick. censored: 'parsnip' Engines for Survival Models. CRAN v0.1.0, 2022.
- M. Krzyziński, M. Spytek, H. Baniecki, P. Biecek. SurvSHAP(t): Time-dependent explanations of machine learning survival models. Knowledge-Based Systems, 2023.