METHYLMAP
Methylmap is a tool for visualization of modified nucleotide frequencies for large cohort sizes.
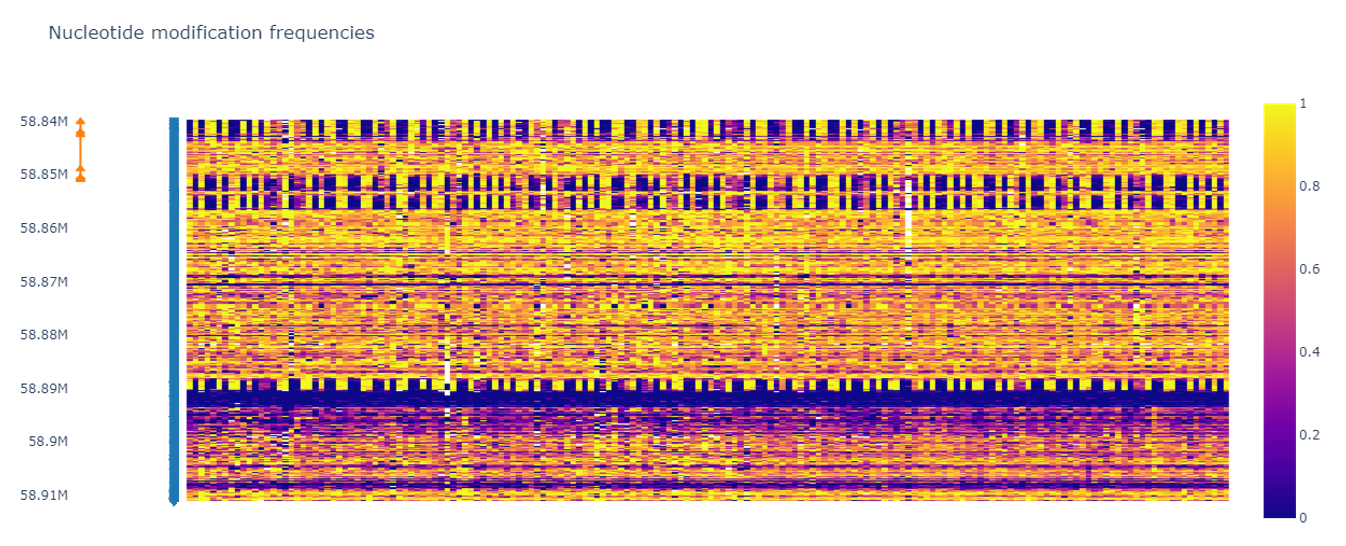
EXAMPLE
INPUT POSSIBILITIES
Supported input possibilities are:
-
BAM/CRAM files with MM and ML tags. Use --files input option.
-
files from nanopolish (as processed by calculate_methylation_frequency.py). The methylation calls can additionally be phased using the available scripts in the "scripts" folder. Use --files input option.
-
an own tab separtated table with nucleotide modification frequencies over all positions (methfreqtable), required header names are "chrom" (column with chromosome information) and "position" (columns with position information). Use --table input option. Example:
chrom position sample_1 sample_2 sample_3 sample_4
0 chr1 100000.0 0.000 0.167 0.000 0.077
1 chr1 100000.5 0.000 0.000 0.100 0.000
2 chr1 100001.0 0.000 0.000 0.000 0.222
3 chr1 100002.0 0.000 0.000 0.000 0.000
4 chr1 100003.0 0.000 0.000 0.000 0.000
- a tab separated file with an overview table containing all nanopolish or BAM/CRAM files and their sample name and experimental group (header requires "path", "name" and "group"). Use --table input option. Example:
path name group
0 /home/path_to_file/bamfile_sample_1.bam samplename_1 case
1 /home/path_to_file/bamfile_sample_2.bam samplename_2 control
2 /home/path_to_file/bamfile_sample_3.bam samplename_3 control
3 /home/path_to_file/bamfile_sample_4.bam samplename_4 case
INSTALLATION
pip install methylmap
USAGE
usage: methylmap [-h] (-f FILES [FILES ...] | -t TABLE) [-w WINDOW] [-n [NAMES ...]] [--gff GFF] [--expand EXPAND]
[--outtable OUTTABLE] [--outfig OUTFIG] [--groups [GROUPS ...]] [-s] [--fasta FASTA]
[--mod {5mC,5hmC,5fC,5caC,5hmU,5fU,5caU,6mA,5oxoG,Xao}] [--dendro] [-v]
Plotting tool for population scale nucleotide modifications.
options:
-h, --help show this help message and exit
-f, --files list with nanopolish (processed with calculate_methylation_frequency.py) files or BAM/CRAM files
-t, --table methfreqtable or overviewtable input
-w, --window region to visualise, format: chr:start-end (example: chr20:58839718-58911192)
-n, --names list with sample names
--gff, --gtf add annotation track based on GTF/GFF file
--expand number of base pairs to expand the window with in both directions
--outtable file to write the frequencies table to in tsv format
--outfig file to write output heatmap to, default: methylmap_{chr}_{start}_{end}.html (missing paths will be created)
--groups list of experimental group for each sample
-s, --simplify simplify annotation track to show genes rather than transcripts
--fasta fasta reference file, required when input is BAM/CRAM files or overviewtable with BAM/CRAM files
--mod modified base of interest when BAM/CRAM files as input. Options are: 5mC, 5hmC, 5fC, 5caC, 5hmU, 5fU, 5caU, 6mA, 5oxoG, Xao, default = 5mC
--dendro perform hierarchical clustering on the samples/haplotypes and visualize with dendrogram on sorted heatmap as output
-v, --version print version and exit
MORE INFORMATION
More information: https://www.biorxiv.org/content/10.1101/2022.11.28.518239v1