This is the official code for our paper:
LENAS: Learning-based Neural Architecture Search and Ensemble for 3D Radiotherapy Dose Prediction
Yi Lin*, Yanfei Liu*, Hao Chen, Xin Yang, Kai Ma, Yefeng Zheng, Kwang-Ting Cheng
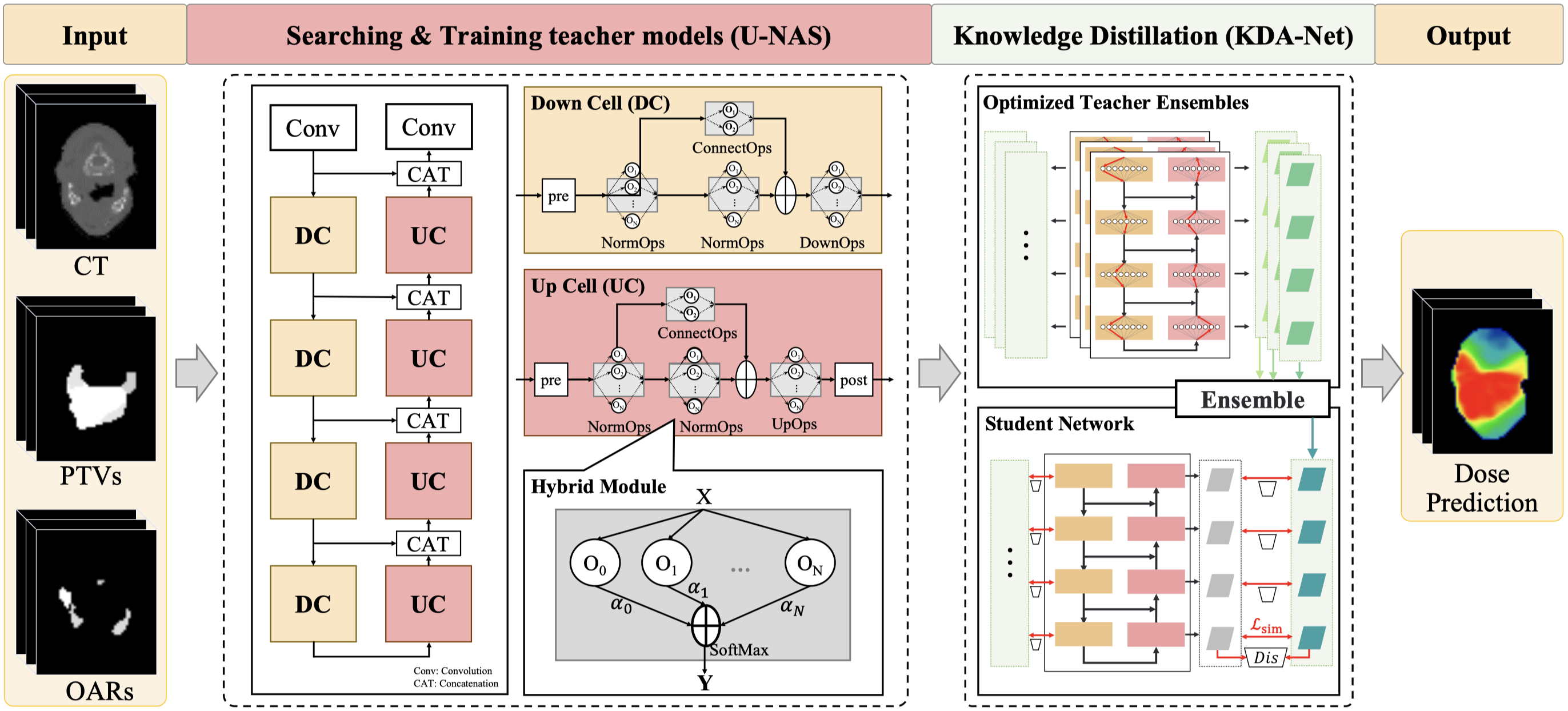
- A learning-based ensemble framework, named LENAS, including the U-NAS framework which efficiently and automatically searches for optimal architectures, and a KDA-Net for the trade-off between the computational cost and accuracy.
- First place in the AIMIS challenge.
pip install -r requirements.txt
- Prepare the data and modify the data path in
config.yml.
cd /path_to_your_RTDosePrediction/RTDosePrediction/DataPrepare
python prepare_OpenKBP_C3D.py
- Search the architecture.
python Main.py
- Training script (Take U-Net as an example).
cd /path_to_your_RTDosePrediction/RTDosePrediction/Unet
python train.py --batch_size 4 --list_GPU_ids 1 0 --max_iter 80000
- Inference script.
cd /path_to_your_RTDosePrediction/RTDosePrediction/Unet
python test.py --GPU_id 0
- The prediction results are stored in
/path_to_your_RTDosePrediction/RTDosePrediction/Output/unet/Prediction.
DataPrepareDataLoaderDataAugmentationNetworkTrainerEvaluate
Teachers: The pre-trained models used as teacher networks.XXX.pklandXXX_geno.pklare the weights and structures, respectively.
- Single Model:
UnetUnet_CBAMVnetFCNDCNN(2D)
- Cascade Models:
C3DC3D_spacing(Resample the data in the data preparation stage)ResC3DResC3D_CBAM
- NAS Manual Model:
Manual_1-Manual_6(See Appendix)
- NAS Single Model:
NAS_18-NAS_42(See Appendix)
- KD Models:
KD
For each MAS Single Model, the best_genotype.pkl file is obtained by manually modifying the geno.py file and running it.
Please cite the paper if you use the code.
TO BE ADDED