Code for papers:
Kanavati, F., Islam, S., Aboagye, E. O., & Rockall, A. (2018). Automatic L3 slice detection in 3D CT images using fully-convolutional networks. arXiv preprint arXiv:1811.09244.
Kanavati, F., Islam, S., Arain, Z., Aboagye, E. O., & Rockall, A. (2020). Fully-automated deep learning slice-based muscle estimation from CT images for sarcopenia assessment. arXiv preprint arXiv:2006.06432.
Trained models for slice detection and slice segmentation are provided in models/
conda create -y --name sarcopenia-ai python=3.6.2
Install
pip install -e .docker build -t sarcopeniaai -f ./Dockerfile .Download the training data from here to your data folder.
docker run --rm -it -v <your_data_folder>:/data -v $(pwd)/configs:/configs sarcopeniaai python -m sarcopenia_ai.apps.slice_detection.trainer --config /configs/slice_detection.cfg
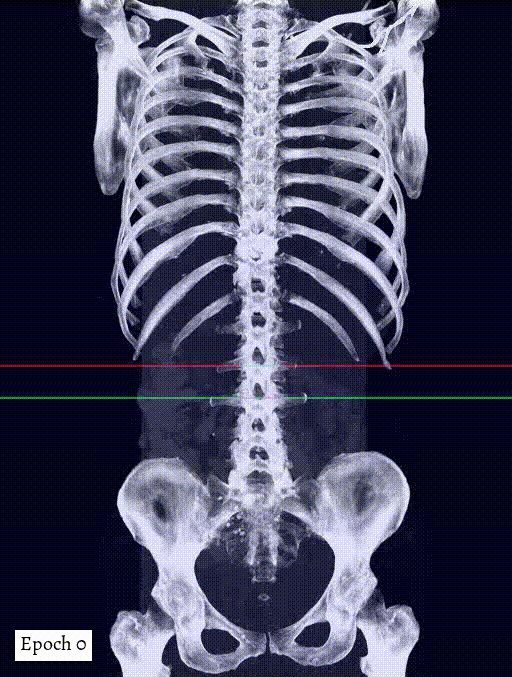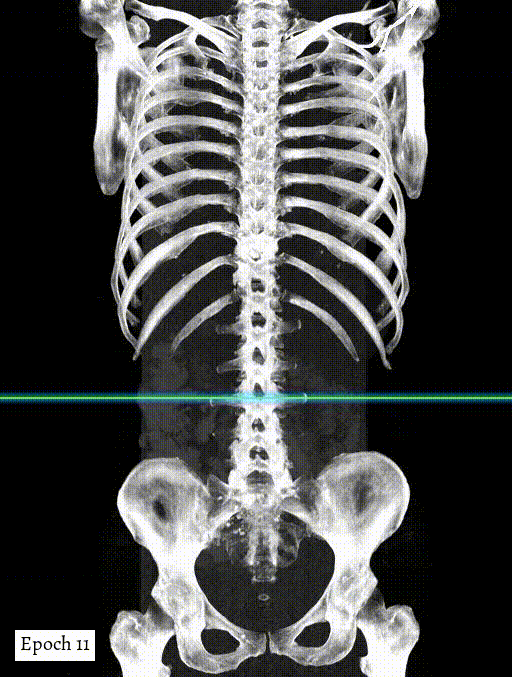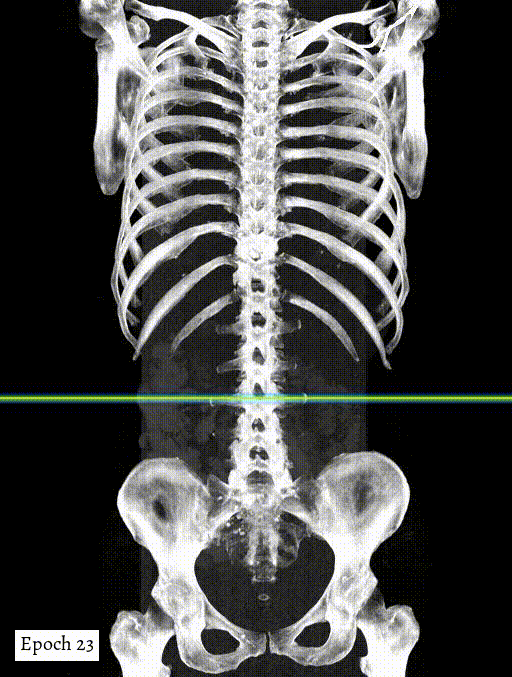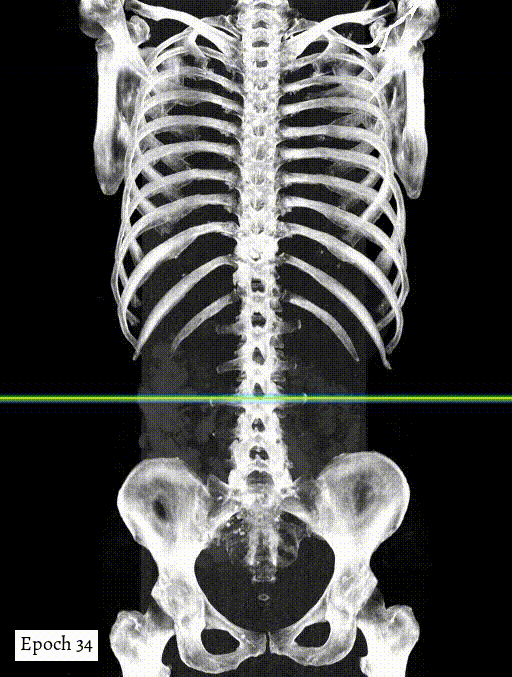
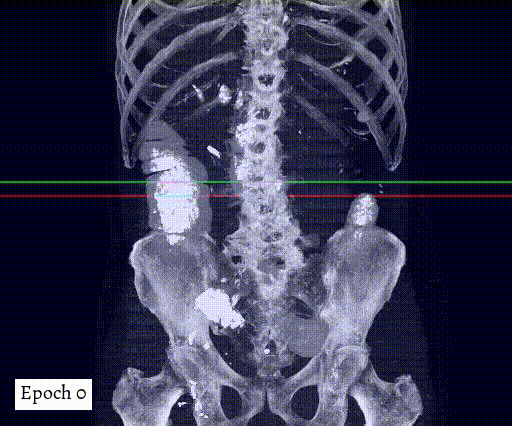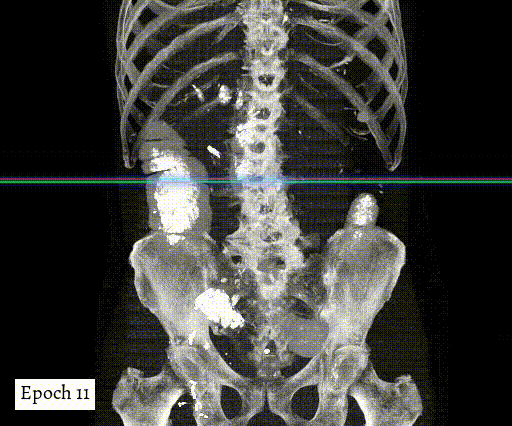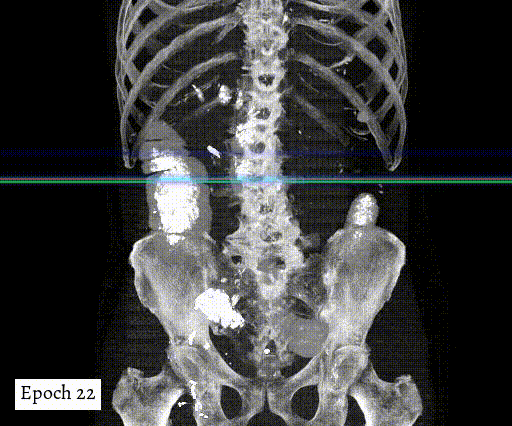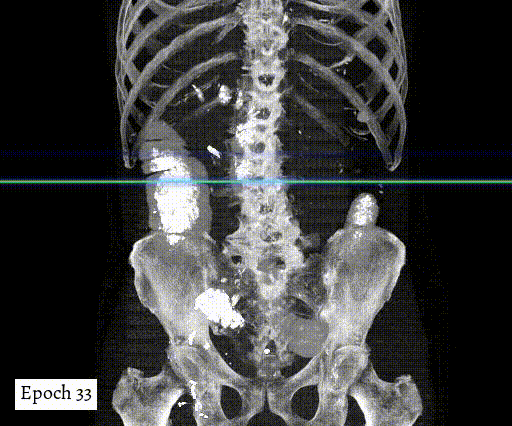
Training output preview on validation images
Labelled segmentation data is not provided. Once you get your own data, you can train a segmentation model with
docker run --rm -it -v <your_data_folder>:/data -v $(pwd)/configs:/configs sarcopeniaai python -m sarcopenia_ai.apps.segmentation.trainer --config /configs/segmentation.cfgdocker run --rm -it -p 5000:5000 sarcopeniaai python -m sarcopenia_ai.apps.server.run_local_server
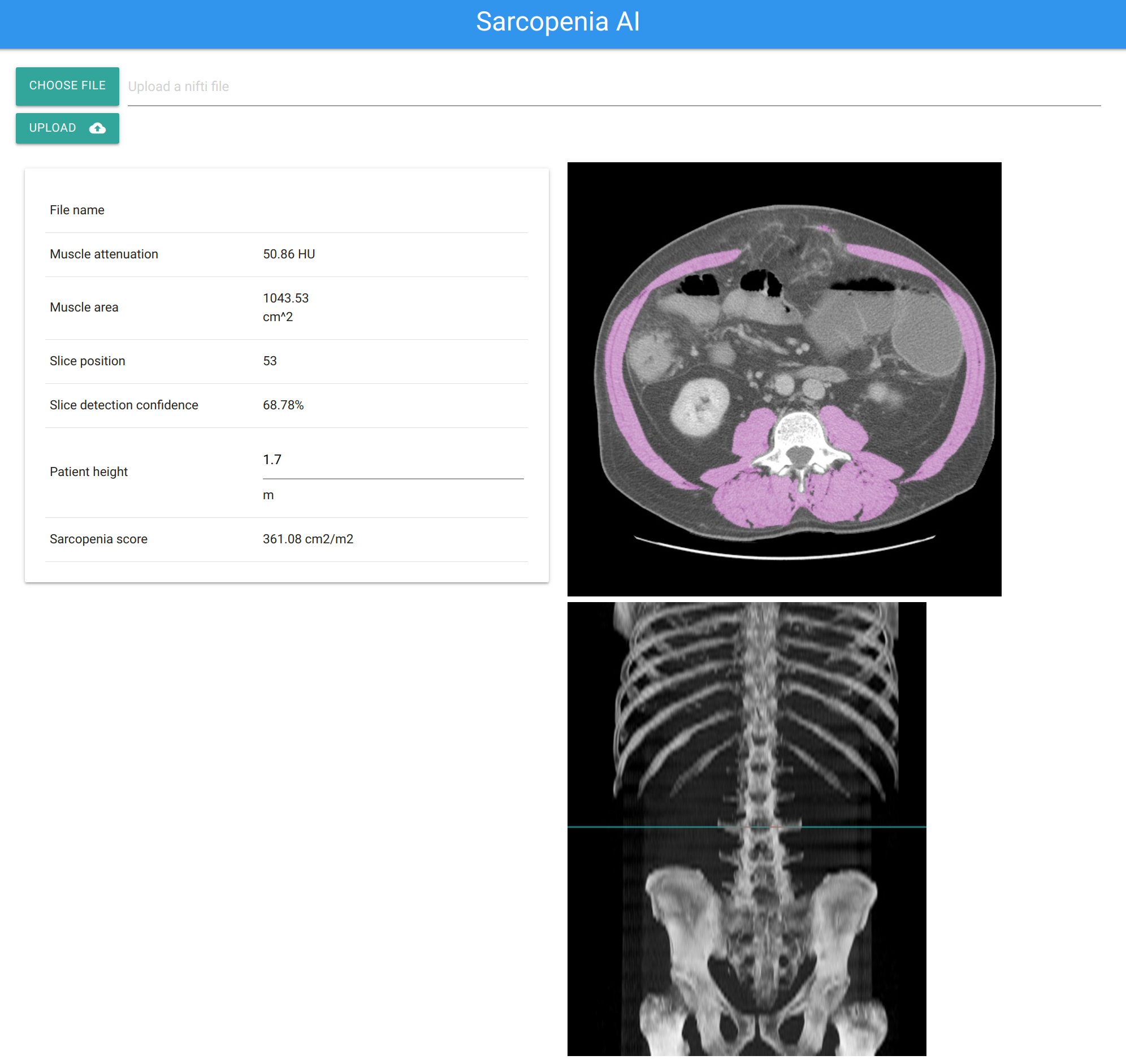
Then head to http://localhost:5000 for web UI
You can also get results from command line. Example:
curl -X POST -F image=@data/volume.nii.gz http://localhost:5000/predict
Expected result
{
"prediction":{
"id":"64667bf3482d4ee5a0e8af6c67b2fa0d",
"muscle_area":"520.15",
"muscle_attenuation":"56.00 HU",
"slice_prob":"69.74%",
"slice_z":90,
"str":"Slice detected at position 90 of 198 with 69.74% confidence "
},
"success":true
}The dataset was collected from multiple sources:
-
3 sets were obtained from the Cancer Imaging Archive (TCIA):
-
a liver tumour dataset was obtained from the LiTS segmentation challenge.
The dataset is available for download in MIPs format from here.
The subset of transitional vertabrae cases can be downloaded from here.
@article{kanavati2018automatic,
title={Automatic L3 slice detection in 3D CT images using fully-convolutional networks},
author={Kanavati, Fahdi and Islam, Shah and Aboagye, Eric O and Rockall, Andrea},
journal={arXiv preprint arXiv:1811.09244},
year={2018}
}
@article{kanavati2020fullyautomated,
title={Fully-automated deep learning slice-based muscle estimation from CT images for sarcopenia assessment},
author={Fahdi Kanavati and Shah Islam and Zohaib Arain and Eric O. Aboagye and Andrea Rockall},
year={2020},
eprint={2006.06432},
archivePrefix={arXiv},
primaryClass={eess.IV}
}