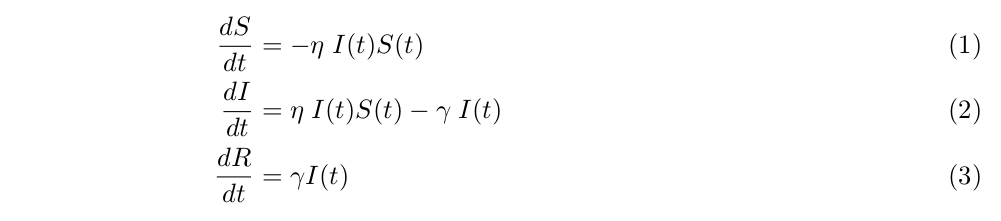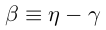| Date | Actual | Predict | Delta | % |
|---|---|---|---|---|
| 2020-02-06 | 31,161 | 34,645 | -3,484 | -11.18 |
| 2020-02-07 | 34,546 | 38,619 | -4,073 | -11.79 |
| 2020-02-08 | 37,198 | 42,490 | -5,292 | -14.23 |
| 2020-02-09 | 40,171 | 45,802 | -5,631 | -14.02 |
| 2020-02-10 | 42,638 | 49,004 | -6,366 | -14.93 |
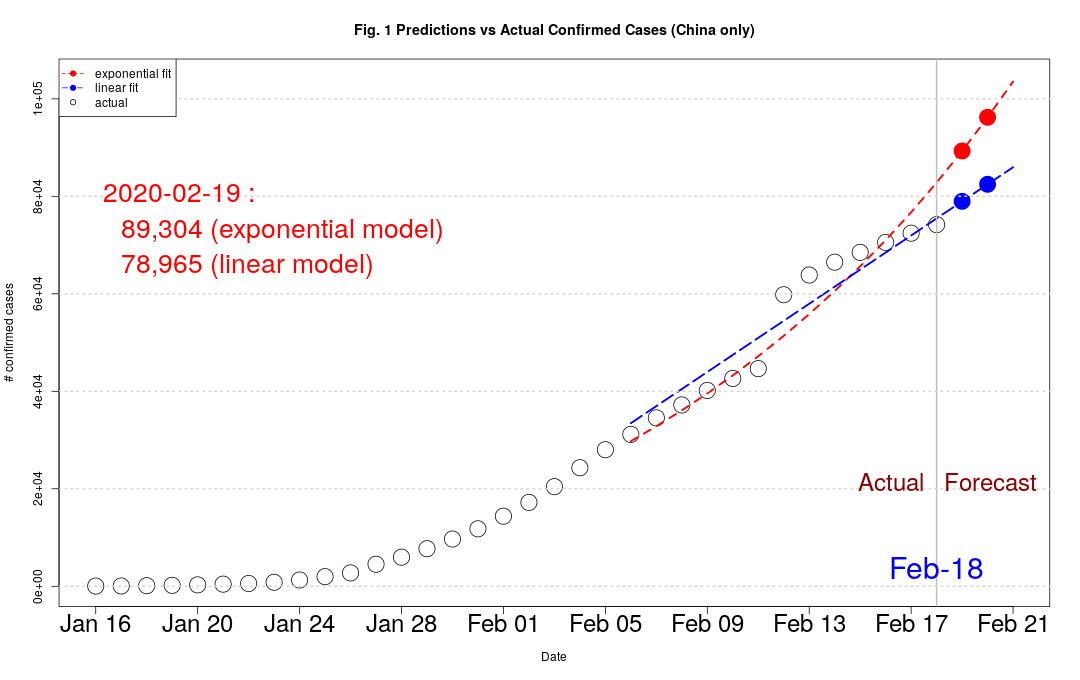
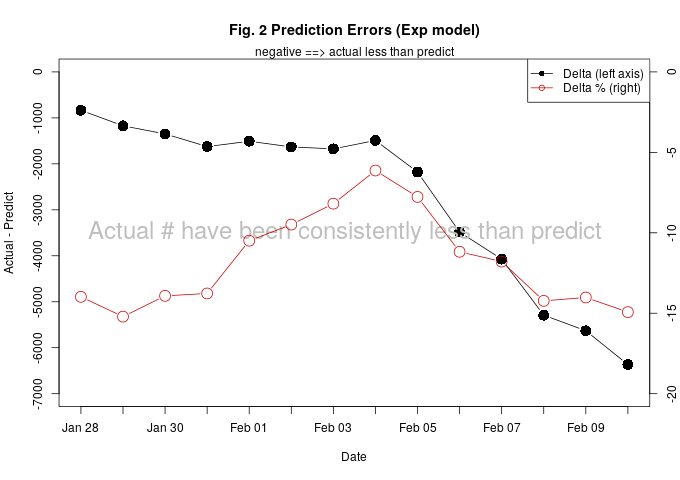
Confirmed cases rising much slower than the exponential model predicted.
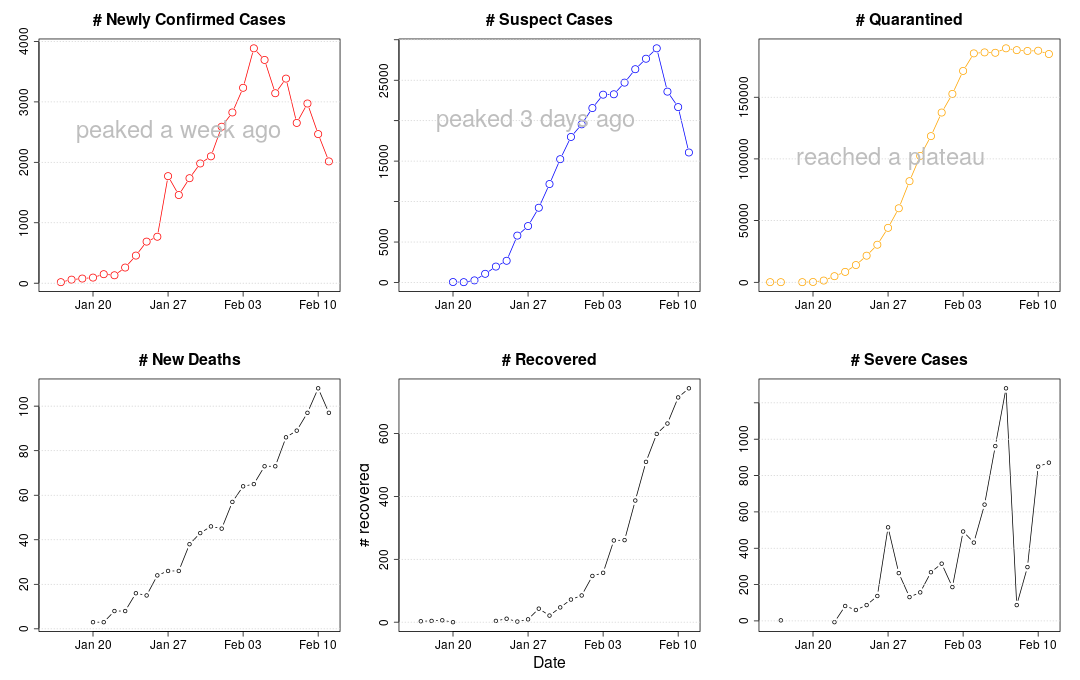
We see a big slowdown in confirmed cases today. Positive.
Observation : Actual has been consistently lower than model prediction. See table below and Fig. 1.
What it means : Spread of the coronavirus is happening at a slower rate than predicted by an exponential model.
Implications : The lock-down in Wuhan and other cities seems to be working.
It's way too early to predict the peak level and its timing, but things don't look as bleak as before.
As the number of infected cases and deaths keep rising, a statistical perspective on the spread of the coronavirus epidemic is needed. This repo presents the modeling tool to make daily prediction. My goal is to make a daily update of the model as new data become available. Epidemiological studies on the 2003 SARS epidemic can be found in [1], [2], and [3]. A discussion of the standard models can be found in [4].
The official source of data is from 中华人民共和国国家卫生健康委员会 (National Health Commission of the PRC) or 卫健委.
http://www.nhc.gov.cn/xcs/yqtb/list_gzbd.shtml
I use the summary table from Wiki. A nice visualization can be found here. I capture the Wiki table and save it in corona_data.csv, which could be read into R.
卫健委 usually updates its website at around US time 16:00 PST.
I use a simplified version of the SIR model, which is a standard formulation of disease transmission dynamics[4]. In a SIR model, we track the time progression of three populations, namely, the susceptible, denoted by the variable S, the infective, by I, and the removed, or R. If a person is healthy and has not contracted the epidemic, he/she belongs to S. A sick person is in I, while R includes both the dead and those who have recovered from and thus are immune to the disease.
The three state variables obey the following coupled non-linear differential equations,
where 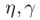 are the parameters of the model, t a discrete time index, and the state variables are fraction of the total population so that
are the parameters of the model, t a discrete time index, and the state variables are fraction of the total population so that 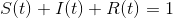 .
.
We could make a simplifying assumption in the case of Wuhan. Since the population in question is large, the impact of the infection is insignificant relative to the size of the population. In a country of 1.4 billion, the current number of deaths from coronavirus is not even 0.01 percent of the population. Therefore, 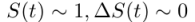 , and
, and 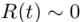 . We could drop Equation (1) and (3), and Equ (2) becomes,
. We could drop Equation (1) and (3), and Equ (2) becomes,
By elementary calculus Equ (4) has solution
The rest is to fit this exponential function to the daily data. I use non-linear least squares in R. For coding details please consult coronavirus.R.
Since the exponential model seems to overestimate the actual number of cases, an alternative is to fit a simple time trend linear model. See coronavirus.R for details.
Fig. 2 shows the prediction errors of the exponential model over time. The prediction errors have been consistently negative. That means the rate of spread has been slower than the best estimate fitted with all the data.
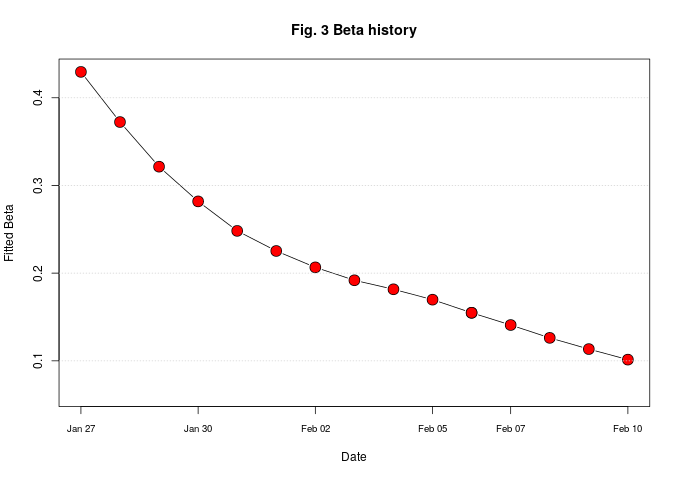
Beta, the rate of infection is dropping, as could be seen in Fig. 3.
Although the number of infected cases continue to rise, the rate of increase has been steadily declining. This is a sign that the lock-down is working.
Open corona_data.csv with your favorite spreadsheet, e.g. Excel. Read off the latest statistics from Wiki and enter them in last row.
Open a terminal,
Rscript coronavirus.R
After the script finishes, two files are generated : latest-prediction.png in the top directory, and Beta-history.png in subdirectory plots/.
To install R, press Ctrl+Alt+T to open a terminal
sudo apt-get update
sudo apt-get install r-base
Code has been tested on
- R 3.6.0
- Ubuntu 18.04
2/11/2020 : R script major rework. Combined exponential and linear model into one plot (latest-prediction.png). Plot functions moved to plot_fun.R which lives in utils\\.
To ask questions or report issues, please open an issue on the issues tracker.
References
[1] Dye, C., & Gay, N. (2003). Modeling the SARS epidemic. Science, 300(5627), 1884-1885.
[2] Brauer, F. (2006). Some simple epidemic models. Mathematical Biosciences & Engineering, 3(1), 1.
[3] Naheed, A., Singh, M., & Lucy, D. (2014). Numerical study of SARS epidemic model with the inclusion of diffusion in the system. Applied Mathematics and computation, 229, 480-498.
[4] Newman, M. E. (2002). Spread of epidemic disease on networks. Physical review E, 66(1), 016128.