- An example of workflow using Docker and GitHub Actions
- This repository gives us some useful techniques such as:
- how to utilize Docker Docker Compose with your Julia workflow.
- how to customize Julia's system image via PackageCompiler.jl to reduce an overhead of package's loading time e.g. Plots.jl, PyCall.jl, or DataFrames.jl etc...
- how to share our work on the Internet. Check our repository on Binder from
- how to use GitHub actions as a CI functionality.
- how to communicate between a Docker container and Juno/Atom
$ tree .
.
├── Dockerfile
├── LICENSE
├── Makefile
├── Project.toml
├── README.md
├── binder
│ └── Dockerfile
├── docker-compose.yml
├── docs
│ ├── Manifest.toml
│ ├── Project.toml
│ ├── make.jl
│ └── src
│ ├── assets
│ │ ├── lab.png
│ │ ├── open_notebook_on_jupyterlab.png
│ │ ├── port9999.png
│ │ └── theorem.css
│ ├── example.md
│ ├── index.md
│ ├── math.md
│ ├── myworkflow.md
│ └── weavesample.jmd
├── experiments
│ └── notebook
│ ├── Chisq.jl
│ ├── Harris.jl
│ ├── Rotation3D.jl
│ ├── apply_PCA_to_MNIST.jl
│ ├── box_and_ball_system.jl
│ ├── coordinate_system.jl
│ ├── curve.jl
│ ├── example.jl
│ ├── fitting.jl
│ ├── gradient_descent.jl
│ ├── histogram_eq.jl
│ ├── hop_step_jump.jl
│ ├── image_filtering.jl
│ ├── interact_sample.jl
│ ├── iris.jl
│ ├── lazysets.jl
│ ├── linear_regression.jl
│ ├── n-Soliton.jl
│ ├── ode.jl
│ ├── plotly_surface.jl
│ ├── plots_sample.jl
│ ├── pyplot.jl
│ ├── seaborn.jl
│ ├── tangent_space.jl
│ └── tangent_vector.jl
├── gitpod
│ └── Dockerfile
├── requirements.txt
├── src
│ └── MyWorkflow.jl
└── test
└── runtests.jl-
Install Docker and Docker Compose. see the following link to learn more with your operating system:
-
To test out you've installed docker, just try:
$ docker run --rm -it julia
# some staff happens ...
- It will initialize the fresh Julia environment even if you do not have a Julia on your (host) machine.
- O.K. Let's build a Docker image for our purpose. Just run:
$ make build
which is exactly equivalent to the following procedure:
$ rm -f Manifest.toml
$ docker build -t myworkflojl .
$ docker-compose build
$ docker-compose run --rm julia julia --project=/work -e 'using Pkg; Pkg.instantiate()'
$ docker-compose up jupyter
myjupyter | To access the notebook, open this file in a browser:
myjupyter | file:///root/.local/share/jupyter/runtime/nbserver-1-open.html
myjupyter | Or copy and paste one of these URLs:
myjupyter | http://4a27c4a06b0f:8888/?token=xxxxxxxxxxxxxxxxxxxxxxx
myjupyter | or http://127.0.0.1:8888/?token=xxxxxxxxxxxxxxxxxxxxxxxThen open your web browser and access to http://127.0.0.1:8888/?token=xxxxxxxxxxxxxxxxxxxxxxx.
You can also initialize JupyterLab as you like via
$ docker-compose up lab- You'll see a JupyterLab screen its theme is Monokai++ (This choice comes from my preference.) :D .
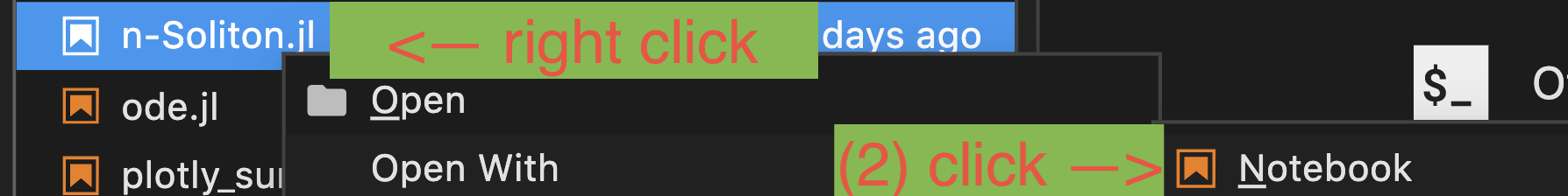
- If you like to open
experiments/notebook/<ournotebook>.jl, right click the file to select andOpen with->Notebook. You're good to go.
-
Note that since we adopt
jupytexttechnology, we do not have to commit/push*.ipynbfile. Namely, we can manage Jupyter Notebook as a normal source code. -
Enjoy your Jupyter life.
Pluto.jl is a lightweight reactive notebooks for Julia. Just run the following command:
$ docker-compose up plutoThen, go to localhost:9999
- We we will assume you've installed Juno.
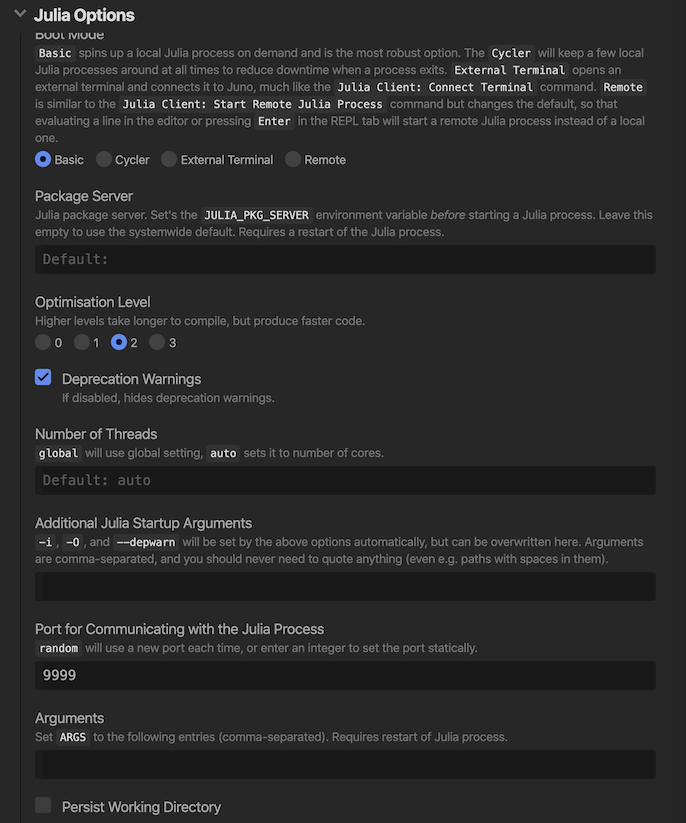
- Go to
Open Settings->Julia Client->Julia Options->Port for Communicating with the Julia processand set value fromrandomto9999.
- To connect to Docker container, open your Atom editor and open command palette(via
Cmd+shift+porCtrl+shift+p). Then selectJulia Client Connect External Process. Finally again open command palette and selectJulia Client: New Terminal. You'll see a terminal at the bottom of the Atom edetor's screen. After that, simply runmake atomor
# For Mac user
$ docker run --rm -it --network=host -v ${PWD}:/work -w /work myworkflowjl julia -J/sysimages/atom.so --project=@. -L .atom/init_mac.jl# For Linux user
$ docker run --rm -it --network=host -v ${PWD}:/work -w /work myworkflowjl julia -J/sysimages/atom.so --project=@. -L .atom/init_linux.jlIt will show Julia's REPL inside of the terminal. pwd() should output "/work", otherwise (e.g. ~/work/MyWorkflow.jl) you're something wrong (opening your Julia session on your host).
julia> pwd()
"/work"- Since our Docker image adopts
sysimagewhich include precompile statements related toAtomorPlots.jlgenerated byPackageCompiler.jl. You'll find the speed ofusing Plots; plot(sin)is much extremely faster than that of runs on Julia session on your host.
# our sysimage
julia> @time begin using Plots; plot(sin) end
0.022140 seconds (38.23 k allocations: 1.731 MiB) # <- Fast
# normal Julia
julia> @time begin using Plots; plot(sin) end
14.006315 seconds (42.16 M allocations: 2.131 GiB, 3.86% gc time) #<- So slow ...
- You can reproduce the
sysimageby yourself to reduce the latency of loading time of heavy packages. See This issue JuliaLang/PackageCompiler.jl#352.